Tapan K. Gandhi
Assistive Completion of Agrammatic Aphasic Sentences: A Transfer Learning Approach using Neurolinguistics-based Synthetic Dataset
Nov 10, 2022Abstract:Damage to the inferior frontal gyrus (Broca's area) can cause agrammatic aphasia wherein patients, although able to comprehend, lack the ability to form complete sentences. This inability leads to communication gaps which cause difficulties in their daily lives. The usage of assistive devices can help in mitigating these issues and enable the patients to communicate effectively. However, due to lack of large scale studies of linguistic deficits in aphasia, research on such assistive technology is relatively limited. In this work, we present two contributions that aim to re-initiate research and development in this field. Firstly, we propose a model that uses linguistic features from small scale studies on aphasia patients and generates large scale datasets of synthetic aphasic utterances from grammatically correct datasets. We show that the mean length of utterance, the noun/verb ratio, and the simple/complex sentence ratio of our synthetic datasets correspond to the reported features of aphasic speech. Further, we demonstrate how the synthetic datasets may be utilized to develop assistive devices for aphasia patients. The pre-trained T5 transformer is fine-tuned using the generated dataset to suggest 5 corrected sentences given an aphasic utterance as input. We evaluate the efficacy of the T5 model using the BLEU and cosine semantic similarity scores. Affirming results with BLEU score of 0.827/1.00 and semantic similarity of 0.904/1.00 were obtained. These results provide a strong foundation for the concept that a synthetic dataset based on small scale studies on aphasia can be used to develop effective assistive technology.
Landmark-Aware and Part-based Ensemble Transfer Learning Network for Facial Expression Recognition from Static images
Apr 22, 2021



Abstract:Facial Expression Recognition from static images is a challenging problem in computer vision applications. Convolutional Neural Network (CNN), the state-of-the-art method for various computer vision tasks, has had limited success in predicting expressions from faces having extreme poses, illumination, and occlusion conditions. To mitigate this issue, CNNs are often accompanied by techniques like transfer, multi-task, or ensemble learning that often provide high accuracy at the cost of high computational complexity. In this work, we propose a Part-based Ensemble Transfer Learning network, which models how humans recognize facial expressions by correlating the spatial orientation pattern of the facial features with a specific expression. It consists of 5 sub-networks, in which each sub-network performs transfer learning from one of the five subsets of facial landmarks: eyebrows, eyes, nose, mouth, or jaw to expression classification. We test the proposed network on the CK+, JAFFE, and SFEW datasets, and it outperforms the benchmark for CK+ and JAFFE datasets by 0.51\% and 5.34\%, respectively. Additionally, it consists of a total of 1.65M model parameters and requires only 3.28 $\times$ $10^{6}$ FLOPS, which ensures computational efficiency for real-time deployment. Grad-CAM visualizations of our proposed ensemble highlight the complementary nature of its sub-networks, a key design parameter of an effective ensemble network. Lastly, cross-dataset evaluation results reveal that our proposed ensemble has a high generalization capacity. Our model trained on the SFEW Train dataset achieves an accuracy of 47.53\% on the CK+ dataset, which is higher than what it achieves on the SFEW Valid dataset.
Identifying Water Stress in Chickpea Plant by Analyzing Progressive Changes in Shoot Images using Deep Learning
Apr 16, 2021



Abstract:To meet the needs of a growing world population, we need to increase the global agricultural yields by employing modern, precision, and automated farming methods. In the recent decade, high-throughput plant phenotyping techniques, which combine non-invasive image analysis and machine learning, have been successfully applied to identify and quantify plant health and diseases. However, these image-based machine learning usually do not consider plant stress's progressive or temporal nature. This time-invariant approach also requires images showing severe signs of stress to ensure high confidence detections, thereby reducing this approach's feasibility for early detection and recovery of plants under stress. In order to overcome the problem mentioned above, we propose a temporal analysis of the visual changes induced in the plant due to stress and apply it for the specific case of water stress identification in Chickpea plant shoot images. For this, we have considered an image dataset of two chickpea varieties JG-62 and Pusa-372, under three water stress conditions; control, young seedling, and before flowering, captured over five months. We then develop an LSTM-CNN architecture to learn visual-temporal patterns from this dataset and predict the water stress category with high confidence. To establish a baseline context, we also conduct a comparative analysis of the CNN architecture used in the proposed model with the other CNN techniques used for the time-invariant classification of water stress. The results reveal that our proposed LSTM-CNN model has resulted in the ceiling level classification performance of \textbf{98.52\%} on JG-62 and \textbf{97.78\%} on Pusa-372 and the chickpea plant data. Lastly, we perform an ablation study to determine the LSTM-CNN model's performance on decreasing the amount of temporal session data used for training.
Performance comparison of 3D correspondence grouping algorithm for 3D plant point clouds
Sep 02, 2019



Abstract:Plant Phenomics can be used to monitor the health and the growth of plants. Computer vision applications like stereo reconstruction, image retrieval, object tracking, and object recognition play an important role in imaging based plant phenotyping. This paper offers a comparative evaluation of some popular 3D correspondence grouping algorithms, motivated by the important role that they can play in tasks such as model creation, plant recognition and identifying plant parts. Another contribution of this paper is the extension of 2D maximum likelihood matching to 3D Maximum Likelihood Estimation Sample Consensus (MLEASAC). MLESAC is efficient and is computationally less intense than 3D random sample consensus (RANSAC). We test these algorithms on 3D point clouds of plants along with two standard benchmarks addressing shape retrieval and point cloud registration scenarios. The performance is evaluated in terms of precision and recall.
Performance Evalution of 3D Keypoint Detectors and Descriptors for Plants Health Classification
Apr 02, 2019



Abstract:Plant Phenomics based on imaging based techniques can be used to monitor the health and the diseases of plants and crops. The use of 3D data for plant phenomics is a recent phenomenon. However, since 3D point cloud contains more information than plant images, in this paper, we compare the performance of different keypoint detectors and local feature descriptors combinations for the plant growth stage and it's growth condition classification based on 3D point clouds of the plants. We have also implemented a modified form of 3D SIFT descriptor, that is invariant to rotation and is computationally less intense than most of the 3D SIFT descriptors reported in the existing literature. The performance is evaluated in terms of the classification accuracy and the results are presented in terms of accuracy tables. We find the ISS-SHOT and the SIFT-SIFT combinations consistently perform better and Fisher Vector (FV) is a better encoder than Vector of Linearly Aggregated (VLAD) for such applications. It can serve as a better modality.
Tumor Classification and Segmentation of MR Brain Images
Oct 31, 2017


Abstract:The diagnosis and segmentation of tumors using any medical diagnostic tool can be challenging due to the varying nature of this pathology. Magnetic Reso- nance Imaging (MRI) is an established diagnostic tool for various diseases and disorders and plays a major role in clinical neuro-diagnosis. Supplementing this technique with automated classification and segmentation tools is gaining importance, to reduce errors and time needed to make a conclusive diagnosis. In this paper a simple three-step algorithm is proposed; (1) identification of patients that present with tumors, (2) automatic selection of abnormal slices of the patients, and (3) segmentation and detection of the tumor. Features were extracted by using discrete wavelet transform on the normalized images and classified by support vector machine (for step (1)) and random forest (for step (2)). The 400 subjects were divided in a 3:1 ratio between training and test with no overlap. This study is novel in terms of use of data, as it employed the entire T2 weighted slices as a single image for classification and a unique combination of contralateral approach with patch thresholding for segmentation, which does not require a training set or a template as is used by most segmentation studies. Using the proposed method, the tumors were segmented accurately with a classification accuracy of 95% with 100% specificity and 90% sensitivity.
Automated Tumor Segmentation and Brain Mapping for the Tumor Area
Oct 28, 2017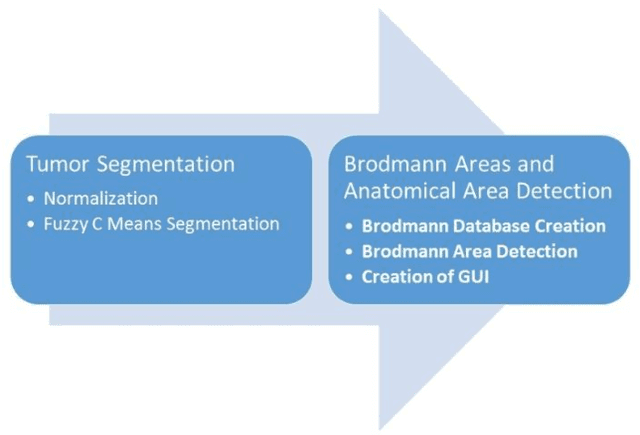
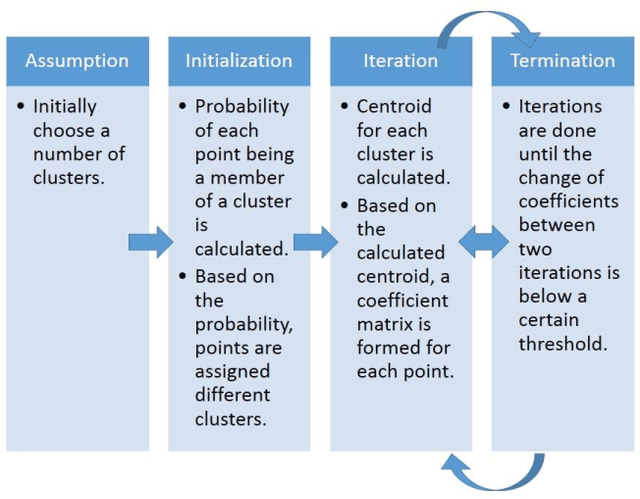
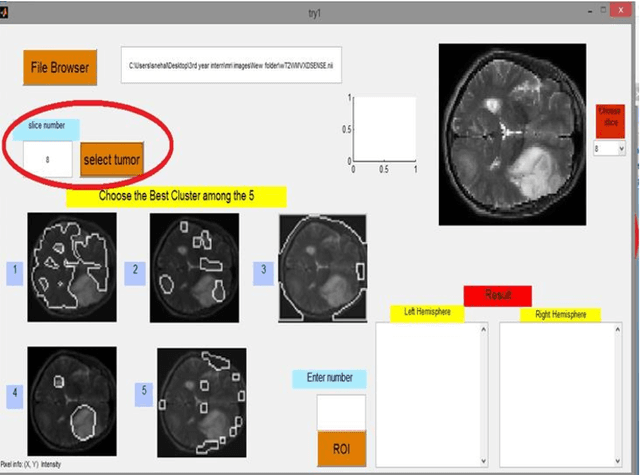

Abstract:Magnetic Resonance Imaging (MRI) is an important diagnostic tool for precise detection of various pathologies. Magnetic Resonance (MR) is more preferred than Computed Tomography (CT) due to the high resolution in MR images which help in better detection of neurological conditions. Graphical user interface (GUI) aided disease detection has become increasingly useful due to the increasing workload of doctors. In this proposed work, a novel two steps GUI technique for brain tumor segmentation as well as Brodmann area detec-tion of the segmented tumor is proposed. A data set of T2 weighted images of 15 patients is used for validating the proposed method. The patient data incor-porates variations in ethnicities, gender (male and female) and age (25-50), thus enhancing the authenticity of the proposed method. The tumors were segmented using Fuzzy C Means Clustering and Brodmann area detection was done using a known template, mapping each area to the segmented tumor image. The proposed method was found to be fairly accurate and robust in detecting tumor.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge