Sylvain Cussat-Blanc
Kartezio: Evolutionary Design of Explainable Pipelines for Biomedical Image Analysis
Feb 28, 2023Abstract:An unresolved issue in contemporary biomedicine is the overwhelming number and diversity of complex images that require annotation, analysis and interpretation. Recent advances in Deep Learning have revolutionized the field of computer vision, creating algorithms that compete with human experts in image segmentation tasks. Crucially however, these frameworks require large human-annotated datasets for training and the resulting models are difficult to interpret. In this study, we introduce Kartezio, a modular Cartesian Genetic Programming based computational strategy that generates transparent and easily interpretable image processing pipelines by iteratively assembling and parameterizing computer vision functions. The pipelines thus generated exhibit comparable precision to state-of-the-art Deep Learning approaches on instance segmentation tasks, while requiring drastically smaller training datasets, a feature which confers tremendous flexibility, speed, and functionality to this approach. We also deployed Kartezio to solve semantic and instance segmentation problems in four real-world Use Cases, and showcase its utility in imaging contexts ranging from high-resolution microscopy to clinical pathology. By successfully implementing Kartezio on a portfolio of images ranging from subcellular structures to tumoral tissue, we demonstrated the flexibility, robustness and practical utility of this fully explicable evolutionary designer for semantic and instance segmentation.
Natural vs Balanced Distribution in Deep Learning on Whole Slide Images for Cancer Detection
Dec 21, 2020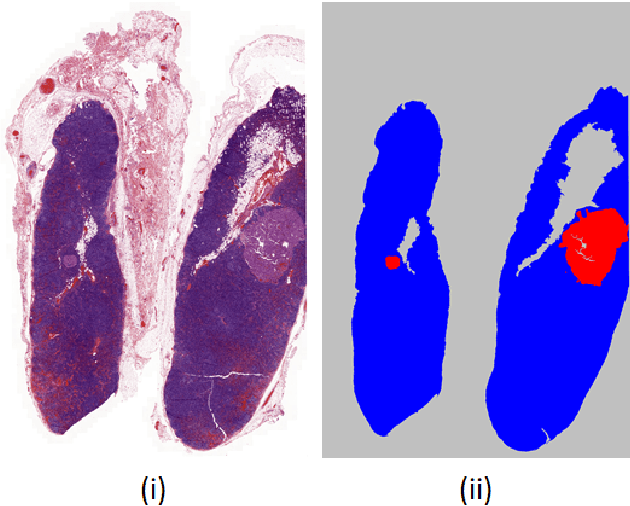
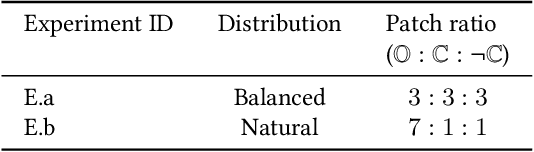
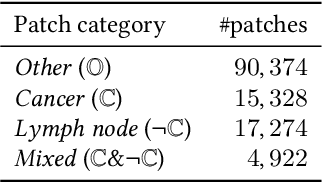
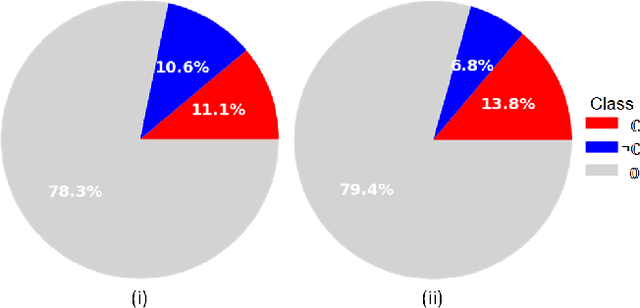
Abstract:The class distribution of data is one of the factors that regulates the performance of machine learning models. However, investigations on the impact of different distributions available in the literature are very few, sometimes absent for domain-specific tasks. In this paper, we analyze the impact of natural and balanced distributions of the training set in deep learning (DL) models applied on histological images, also known as whole slide images (WSIs). WSIs are considered as the gold standard for cancer diagnosis. In recent years, researchers have turned their attention to DL models to automate and accelerate the diagnosis process. In the training of such DL models, filtering out the non-regions-of-interest from the WSIs and adopting an artificial distribution (usually, a balanced distribution) is a common trend. In our analysis, we show that keeping the WSIs data in their usual distribution (which we call natural distribution) for DL training produces fewer false positives (FPs) with comparable false negatives (FNs) than the artificially-obtained balanced distribution. We conduct an empirical comparative study with 10 random folds for each distribution, comparing the resulting average performance levels in terms of five different evaluation metrics. Experimental results show the effectiveness of the natural distribution over the balanced one across all the evaluation metrics.
Neuromodulated Learning in Deep Neural Networks
Dec 05, 2018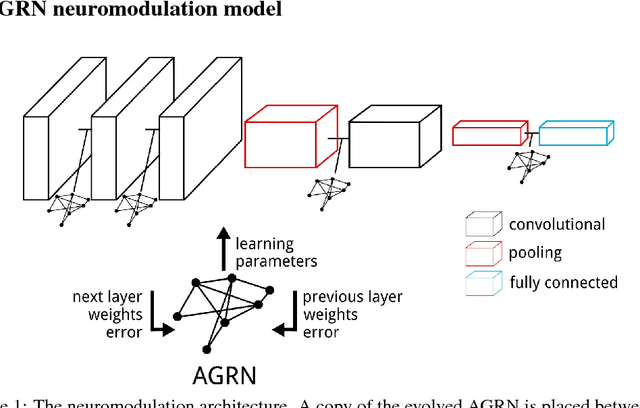
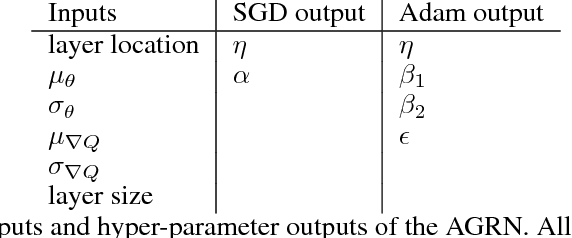

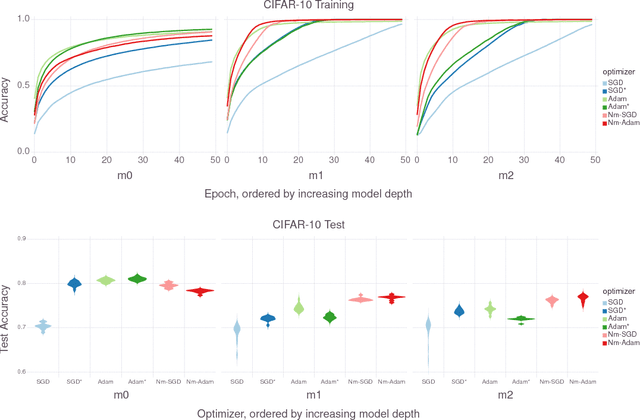
Abstract:In the brain, learning signals change over time and synaptic location, and are applied based on the learning history at the synapse, in the complex process of neuromodulation. Learning in artificial neural networks, on the other hand, is shaped by hyper-parameters set before learning starts, which remain static throughout learning, and which are uniform for the entire network. In this work, we propose a method of deep artificial neuromodulation which applies the concepts of biological neuromodulation to stochastic gradient descent. Evolved neuromodulatory dynamics modify learning parameters at each layer in a deep neural network over the course of the network's training. We show that the same neuromodulatory dynamics can be applied to different models and can scale to new problems not encountered during evolution. Finally, we examine the evolved neuromodulation, showing that evolution found dynamic, location-specific learning strategies.
Positional Cartesian Genetic Programming
Oct 09, 2018



Abstract:Cartesian Genetic Programming (CGP) has many modifications across a variety of implementations, such as recursive connections and node weights. Alternative genetic operators have also been proposed for CGP, but have not been fully studied. In this work, we present a new form of genetic programming based on a floating point representation. In this new form of CGP, called Positional CGP, node positions are evolved. This allows for the evaluation of many different genetic operators while allowing for previous CGP improvements like recurrency. Using nine benchmark problems from three different classes, we evaluate the optimal parameters for CGP and PCGP, including novel genetic operators.
Evolving Differentiable Gene Regulatory Networks
Jul 16, 2018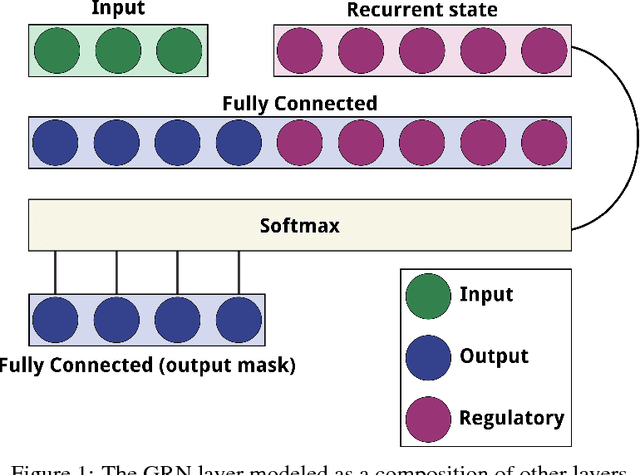
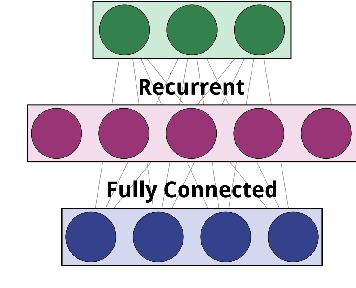
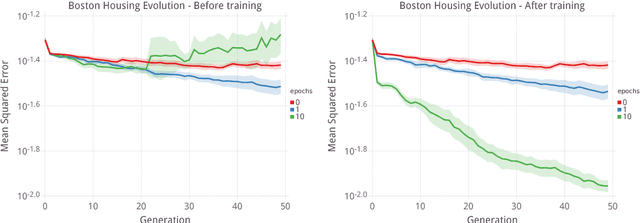
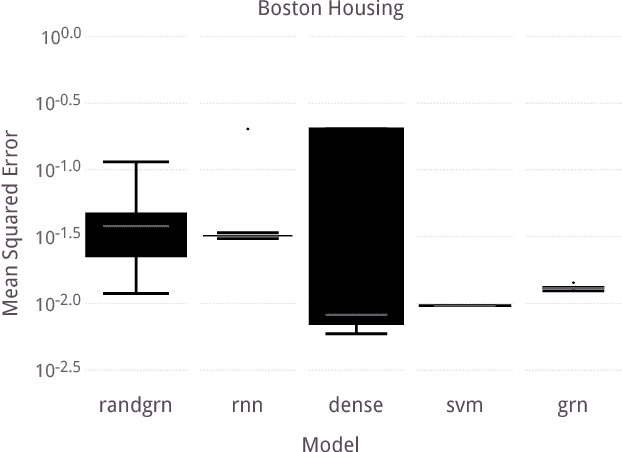
Abstract:Over the past twenty years, artificial Gene Regulatory Networks (GRNs) have shown their capacity to solve real-world problems in various domains such as agent control, signal processing and artificial life experiments. They have also benefited from new evolutionary approaches and improvements to dynamic which have increased their optimization efficiency. In this paper, we present an additional step toward their usability in machine learning applications. We detail an GPU-based implementation of differentiable GRNs, allowing for local optimization of GRN architectures with stochastic gradient descent (SGD). Using a standard machine learning dataset, we evaluate the ways in which evolution and SGD can be combined to further GRN optimization. We compare these approaches with neural network models trained by SGD and with support vector machines.
Evolving simple programs for playing Atari games
Jun 14, 2018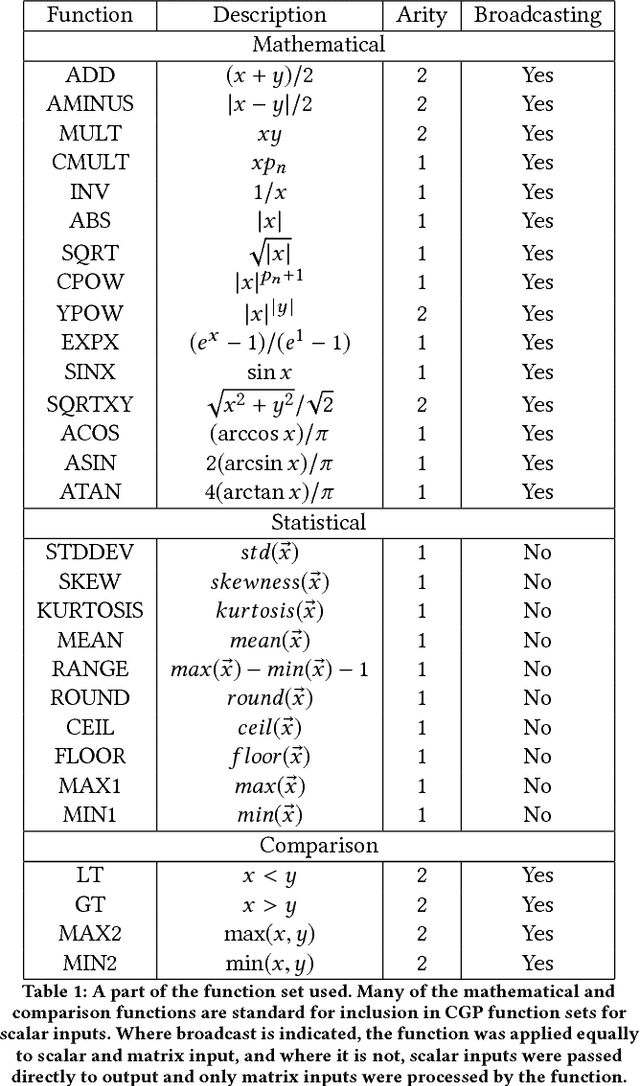
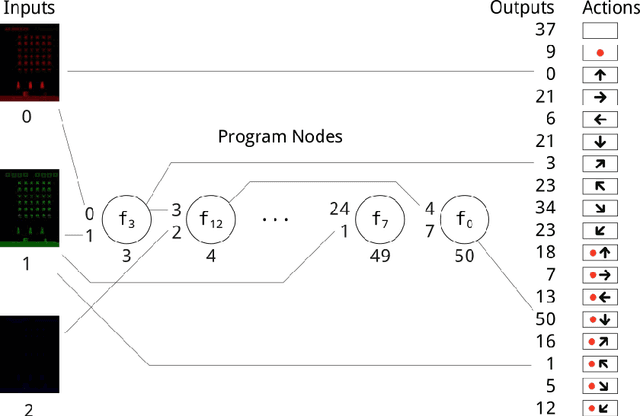

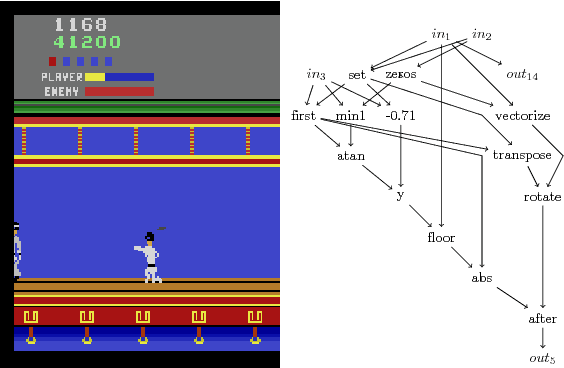
Abstract:Cartesian Genetic Programming (CGP) has previously shown capabilities in image processing tasks by evolving programs with a function set specialized for computer vision. A similar approach can be applied to Atari playing. Programs are evolved using mixed type CGP with a function set suited for matrix operations, including image processing, but allowing for controller behavior to emerge. While the programs are relatively small, many controllers are competitive with state of the art methods for the Atari benchmark set and require less training time. By evaluating the programs of the best evolved individuals, simple but effective strategies can be found.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge