Susanna Dodd
Assessment of contextualised representations in detecting outcome phrases in clinical trials
Mar 13, 2022



Abstract:Automating the recognition of outcomes reported in clinical trials using machine learning has a huge potential of speeding up access to evidence necessary in healthcare decision-making. Prior research has however acknowledged inadequate training corpora as a challenge for the Outcome detection (OD) task. Additionally, several contextualized representations like BERT and ELMO have achieved unparalleled success in detecting various diseases, genes, proteins, and chemicals, however, the same cannot be emphatically stated for outcomes, because these models have been relatively under-tested and studied for the OD task. We introduce "EBM-COMET", a dataset in which 300 PubMed abstracts are expertly annotated for clinical outcomes. Unlike prior related datasets that use arbitrary outcome classifications, we use labels from a taxonomy recently published to standardize outcome classifications. To extract outcomes, we fine-tune a variety of pre-trained contextualized representations, additionally, we use frozen contextualized and context-independent representations in our custom neural model augmented with clinically informed Part-Of-Speech embeddings and a cost-sensitive loss function. We adopt strict evaluation for the trained models by rewarding them for correctly identifying full outcome phrases rather than words within the entities i.e. given an outcome "systolic blood pressure", the models are rewarded a classification score only when they predict all 3 words in sequence, otherwise, they are not rewarded. We observe our best model (BioBERT) achieve 81.5\% F1, 81.3\% sensitivity and 98.0\% specificity. We reach a consensus on which contextualized representations are best suited for detecting outcomes from clinical-trial abstracts. Furthermore, our best model outperforms scores published on the original EBM-NLP dataset leader-board scores.
Detect and Classify -- Joint Span Detection and Classification for Health Outcomes
Apr 15, 2021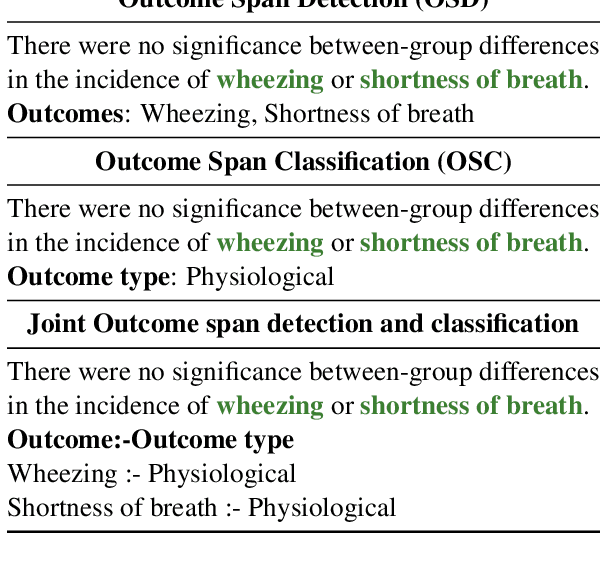
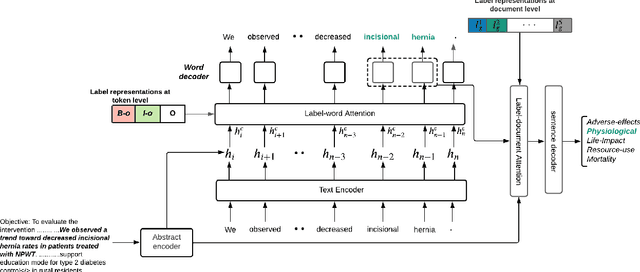
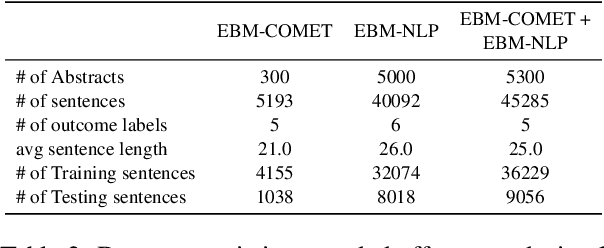
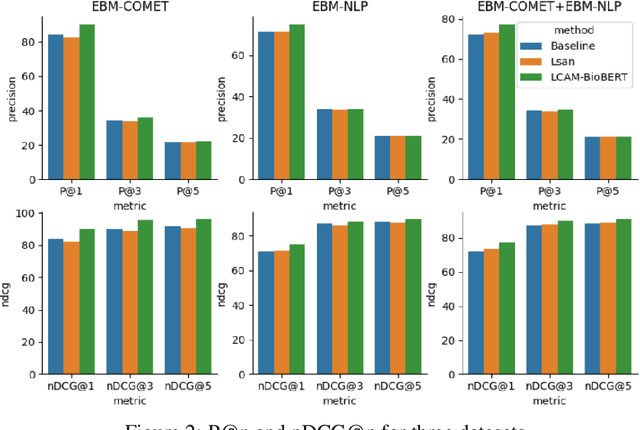
Abstract:A health outcome is a measurement or an observation used to capture and assess the effect of a treatment. Automatic detection of health outcomes from text would undoubtedly speed up access to evidence necessary in healthcare decision making. Prior work on outcome detection has modelled this task as either (a) a sequence labelling task, where the goal is to detect which text spans describe health outcomes or (b) a classification task, where the goal is to classify a text into a pre-defined set of categories depending on an outcome that is mentioned somewhere in that text. However, this decoupling of span detection and classification is problematic from a modelling perspective and ignores global structural correspondences between sentence-level and word-level information present in a given text. We propose a method that uses both word-level and sentence-level information to simultaneously perform outcome span detection and outcome type classification. In addition to injecting contextual information to hidden vectors, we use label attention to appropriately weight both word-level and sentence-level information. Experimental results on several benchmark datasets for health outcome detection show that our model consistently outperforms decoupled methods, reporting competitive results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge