Suraj Pai
Artificial Intelligence in Medicine, Radiology and Nuclear Medicine, CARIM & GROW, Maastricht University, Department of Radiation Oncology, Brigham and Women's Hospital, Dana-Farber Cancer Institute, Harvard Medical School
MHub.ai: A Simple, Standardized, and Reproducible Platform for AI Models in Medical Imaging
Jan 15, 2026Abstract:Artificial intelligence (AI) has the potential to transform medical imaging by automating image analysis and accelerating clinical research. However, research and clinical use are limited by the wide variety of AI implementations and architectures, inconsistent documentation, and reproducibility issues. Here, we introduce MHub.ai, an open-source, container-based platform that standardizes access to AI models with minimal configuration, promoting accessibility and reproducibility in medical imaging. MHub.ai packages models from peer-reviewed publications into standardized containers that support direct processing of DICOM and other formats, provide a unified application interface, and embed structured metadata. Each model is accompanied by publicly available reference data that can be used to confirm model operation. MHub.ai includes an initial set of state-of-the-art segmentation, prediction, and feature extraction models for different modalities. The modular framework enables adaptation of any model and supports community contributions. We demonstrate the utility of the platform in a clinical use case through comparative evaluation of lung segmentation models. To further strengthen transparency and reproducibility, we publicly release the generated segmentations and evaluation metrics and provide interactive dashboards that allow readers to inspect individual cases and reproduce or extend our analysis. By simplifying model use, MHub.ai enables side-by-side benchmarking with identical execution commands and standardized outputs, and lowers the barrier to clinical translation.
Foundation Artificial Intelligence Models for Health Recognition Using Face Photographs (FAHR-Face)
Jun 17, 2025Abstract:Background: Facial appearance offers a noninvasive window into health. We built FAHR-Face, a foundation model trained on >40 million facial images and fine-tuned it for two distinct tasks: biological age estimation (FAHR-FaceAge) and survival risk prediction (FAHR-FaceSurvival). Methods: FAHR-FaceAge underwent a two-stage, age-balanced fine-tuning on 749,935 public images; FAHR-FaceSurvival was fine-tuned on 34,389 photos of cancer patients. Model robustness (cosmetic surgery, makeup, pose, lighting) and independence (saliency mapping) was tested extensively. Both models were clinically tested in two independent cancer patient datasets with survival analyzed by multivariable Cox models and adjusted for clinical prognostic factors. Findings: For age estimation, FAHR-FaceAge had the lowest mean absolute error of 5.1 years on public datasets, outperforming benchmark models and maintaining accuracy across the full human lifespan. In cancer patients, FAHR-FaceAge outperformed a prior facial age estimation model in survival prognostication. FAHR-FaceSurvival demonstrated robust prediction of mortality, and the highest-risk quartile had more than triple the mortality of the lowest (adjusted hazard ratio 3.22; P<0.001). These findings were validated in the independent cohort and both models showed generalizability across age, sex, race and cancer subgroups. The two algorithms provided distinct, complementary prognostic information; saliency mapping revealed each model relied on distinct facial regions. The combination of FAHR-FaceAge and FAHR-FaceSurvival improved prognostic accuracy. Interpretation: A single foundation model can generate inexpensive, scalable facial biomarkers that capture both biological ageing and disease-related mortality risk. The foundation model enabled effective training using relatively small clinical datasets.
Vision Foundation Models for Computed Tomography
Jan 15, 2025Abstract:Foundation models (FMs) have shown transformative potential in radiology by performing diverse, complex tasks across imaging modalities. Here, we developed CT-FM, a large-scale 3D image-based pre-trained model designed explicitly for various radiological tasks. CT-FM was pre-trained using 148,000 computed tomography (CT) scans from the Imaging Data Commons through label-agnostic contrastive learning. We evaluated CT-FM across four categories of tasks, namely, whole-body and tumor segmentation, head CT triage, medical image retrieval, and semantic understanding, showing superior performance against state-of-the-art models. Beyond quantitative success, CT-FM demonstrated the ability to cluster regions anatomically and identify similar anatomical and structural concepts across scans. Furthermore, it remained robust across test-retest settings and indicated reasonable salient regions attached to its embeddings. This study demonstrates the value of large-scale medical imaging foundation models and by open-sourcing the model weights, code, and data, aims to support more adaptable, reliable, and interpretable AI solutions in radiology.
Exploring constraints on CycleGAN-based CBCT enhancement for adaptive radiotherapy
Oct 12, 2021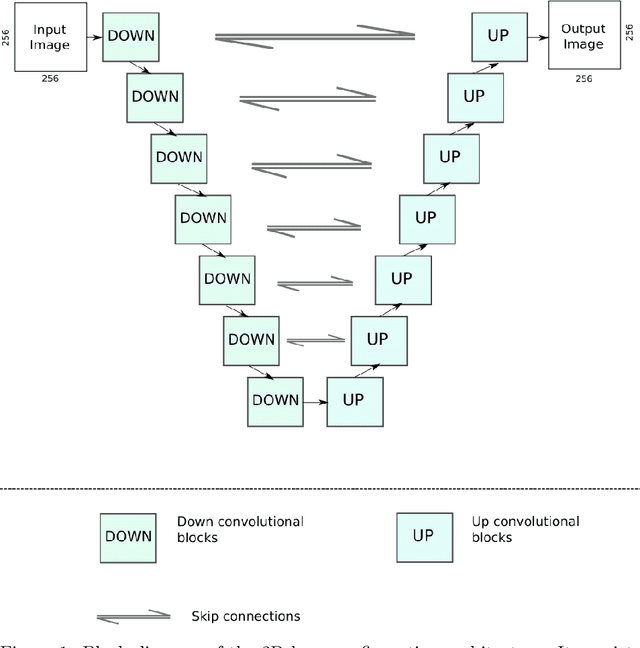



Abstract:Research exploring CycleGAN-based synthetic image generation has recently accelerated in the medical community, as it is able to leverage unpaired datasets effectively. However, clinical acceptance of these synthetic images pose a significant challenge as they are subject to strict evaluation protocols. A commonly established drawback of the CycleGAN, the introduction of artifacts in generated images is unforgivable in the case of medical images. In an attempt to alleviate this drawback, we explore different constraints of the CycleGAN along with investigation of adaptive control of these constraints. The benefits of imposing additional constraints on the CycleGAN, in the form of structure retaining losses is also explored. A generalized frequency loss inspired by arxiv:2012.12821 that preserves content in the frequency domain between source and target is investigated and compared with existing losses such as the MIND loss arXiv:1809.04536. CycleGAN implementations from the ganslate framework (https://github.com/ganslate-team/ganslate) are used for experimentation in this thesis. Synthetic images generated from our methods are quantitatively and qualitatively investigated and outperform the baseline CycleGAN and other approaches. Furthermore, no observable artifacts or loss in image quality is found, which is critical for acceptance of these synthetic images. The synthetic medical images thus generated are also evaluated using domain-specific evaluation and using segmentation as a downstream task, in order to clearly highlight their applicability to clinical workflows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge