Sungkyu Jung
Subspace Recovery in Winsorized PCA: Insights into Accuracy and Robustness
Feb 23, 2025Abstract:In this paper, we explore the theoretical properties of subspace recovery using Winsorized Principal Component Analysis (WPCA), utilizing a common data transformation technique that caps extreme values to mitigate the impact of outliers. Despite the widespread use of winsorization in various tasks of multivariate analysis, its theoretical properties, particularly for subspace recovery, have received limited attention. We provide a detailed analysis of the accuracy of WPCA, showing that increasing the number of samples while decreasing the proportion of outliers guarantees the consistency of the sample subspaces from WPCA with respect to the true population subspace. Furthermore, we establish perturbation bounds that ensure the WPCA subspace obtained from contaminated data remains close to the subspace recovered from pure data. Additionally, we extend the classical notion of breakdown points to subspace-valued statistics and derive lower bounds for the breakdown points of WPCA. Our analysis demonstrates that WPCA exhibits strong robustness to outliers while maintaining consistency under mild assumptions. A toy example is provided to numerically illustrate the behavior of the upper bounds for perturbation bounds and breakdown points, emphasizing winsorization's utility in subspace recovery.
Robust SVD Made Easy: A fast and reliable algorithm for large-scale data analysis
Feb 15, 2024Abstract:The singular value decomposition (SVD) is a crucial tool in machine learning and statistical data analysis. However, it is highly susceptible to outliers in the data matrix. Existing robust SVD algorithms often sacrifice speed for robustness or fail in the presence of only a few outliers. This study introduces an efficient algorithm, called Spherically Normalized SVD, for robust SVD approximation that is highly insensitive to outliers, computationally scalable, and provides accurate approximations of singular vectors. The proposed algorithm achieves remarkable speed by utilizing only two applications of a standard reduced-rank SVD algorithm to appropriately scaled data, significantly outperforming competing algorithms in computation times. To assess the robustness of the approximated singular vectors and their subspaces against data contamination, we introduce new notions of breakdown points for matrix-valued input, including row-wise, column-wise, and block-wise breakdown points. Theoretical and empirical analyses demonstrate that our algorithm exhibits higher breakdown points compared to standard SVD and its modifications. We empirically validate the effectiveness of our approach in applications such as robust low-rank approximation and robust principal component analysis of high-dimensional microarray datasets. Overall, our study presents a highly efficient and robust solution for SVD approximation that overcomes the limitations of existing algorithms in the presence of outliers.
Covariance-engaged Classification of Sets via Linear Programming
Jun 26, 2020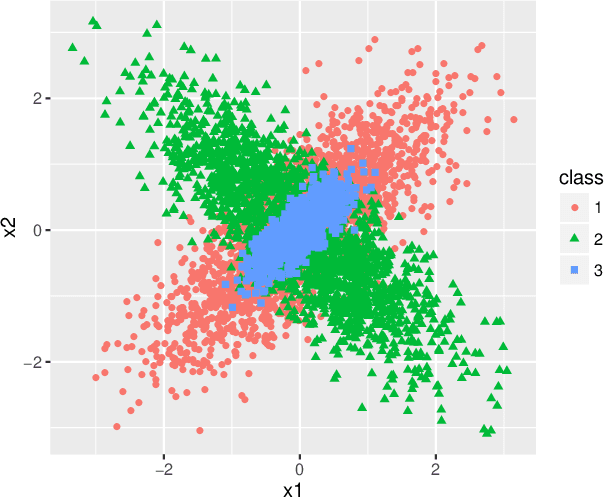
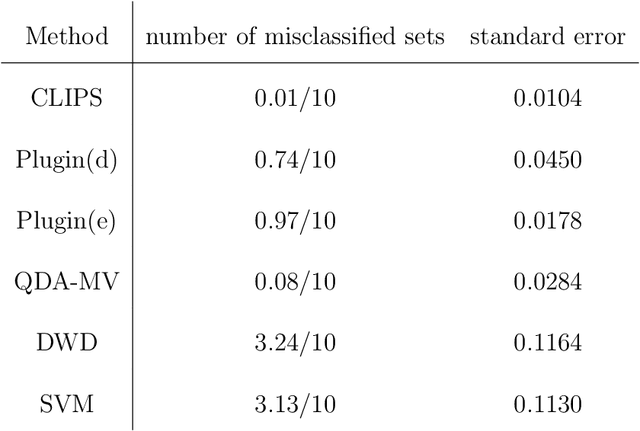
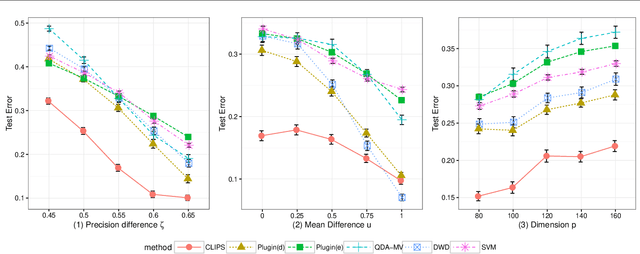
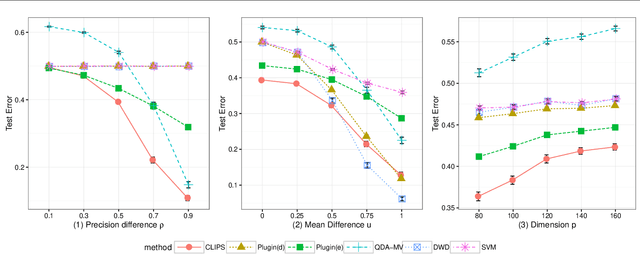
Abstract:Set classification aims to classify a set of observations as a whole, as opposed to classifying individual observations separately. To formally understand the unfamiliar concept of binary set classification, we first investigate the optimal decision rule under the normal distribution, which utilizes the empirical covariance of the set to be classified. We show that the number of observations in the set plays a critical role in bounding the Bayes risk. Under this framework, we further propose new methods of set classification. For the case where only a few parameters of the model drive the difference between two classes, we propose a computationally-efficient approach to parameter estimation using linear programming, leading to the Covariance-engaged LInear Programming Set (CLIPS) classifier. Its theoretical properties are investigated for both independent case and various (short-range and long-range dependent) time series structures among observations within each set. The convergence rates of estimation errors and risk of the CLIPS classifier are established to show that having multiple observations in a set leads to faster convergence rates, compared to the standard classification situation in which there is only one observation in the set. The applicable domains in which the CLIPS performs better than competitors are highlighted in a comprehensive simulation study. Finally, we illustrate the usefulness of the proposed methods in classification of real image data in histopathology.
Continuum directions for supervised dimension reduction
Mar 21, 2018



Abstract:Dimension reduction of multivariate data supervised by auxiliary information is considered. A series of basis for dimension reduction is obtained as minimizers of a novel criterion. The proposed method is akin to continuum regression, and the resulting basis is called continuum directions. With a presence of binary supervision data, these directions continuously bridge the principal component, mean difference and linear discriminant directions, thus ranging from unsupervised to fully supervised dimension reduction. High-dimensional asymptotic studies of continuum directions for binary supervision reveal several interesting facts. The conditions under which the sample continuum directions are inconsistent, but their classification performance is good, are specified. While the proposed method can be directly used for binary and multi-category classification, its generalizations to incorporate any form of auxiliary data are also presented. The proposed method enjoys fast computation, and the performance is better or on par with more computer-intensive alternatives.
A Statistical Approach to Set Classification by Feature Selection with Applications to Classification of Histopathology Images
Feb 19, 2014



Abstract:Set classification problems arise when classification tasks are based on sets of observations as opposed to individual observations. In set classification, a classification rule is trained with $N$ sets of observations, where each set is labeled with class information, and the prediction of a class label is performed also with a set of observations. Data sets for set classification appear, for example, in diagnostics of disease based on multiple cell nucleus images from a single tissue. Relevant statistical models for set classification are introduced, which motivate a set classification framework based on context-free feature extraction. By understanding a set of observations as an empirical distribution, we employ a data-driven method to choose those features which contain information on location and major variation. In particular, the method of principal component analysis is used to extract the features of major variation. Multidimensional scaling is used to represent features as vector-valued points on which conventional classifiers can be applied. The proposed set classification approaches achieve better classification results than competing methods in a number of simulated data examples. The benefits of our method are demonstrated in an analysis of histopathology images of cell nuclei related to liver cancer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge