Steve Shirtliffe
The Normalized Difference Layer: A Differentiable Spectral Index Formulation for Deep Learning
Jan 11, 2026Abstract:Normalized difference indices have been a staple in remote sensing for decades. They stay reliable under lighting changes produce bounded values and connect well to biophysical signals. Even so, they are usually treated as a fixed pre processing step with coefficients set to one, which limits how well they can adapt to a specific learning task. In this study, we introduce the Normalized Difference Layer that is a differentiable neural network module. The proposed method keeps the classical idea but learns the band coefficients from data. We present a complete mathematical framework for integrating this layer into deep learning architectures that uses softplus reparameterization to ensure positive coefficients and bounded denominators. We describe forward and backward pass algorithms enabling end to end training through backpropagation. This approach preserves the key benefits of normalized differences, namely illumination invariance and outputs bounded to $[-1,1]$ while allowing gradient descent to discover task specific band weightings. We extend the method to work with signed inputs, so the layer can be stacked inside larger architectures. Experiments show that models using this layer reach similar classification accuracy to standard multilayer perceptrons while using about 75\% fewer parameters. They also handle multiplicative noise well, at 10\% noise accuracy drops only 0.17\% versus 3.03\% for baseline MLPs. The learned coefficient patterns stay consistent across different depths.
Automated Discovery of Parsimonious Spectral Indices via Normalized Difference Polynomials
Dec 26, 2025Abstract:We introduce an automated way to find compact spectral indices for vegetation classification. The idea is to take all pairwise normalized differences from the spectral bands and then build polynomial combinations up to a fixed degree, which gives a structured search space that still keeps the illumination invariance needed in remote sensing. For a sensor with $n$ bands this produces $\binom{n}{2}$ base normalized differences, and the degree-2 polynomial expansion gives 1,080 candidate features for the 10-band Sentinel-2 configuration we use here. Feature selection methods (ANOVA filtering, recursive elimination, and $L_1$-regularized SVM) then pick out small sets of indices that reach the desired accuracy, so the final models stay simple and easy to interpret. We test the framework on Kochia (\textit{Bassia scoparia}) detection using Sentinel-2 imagery from Saskatchewan, Canada ($N = 2{,}318$ samples, 2022--2024). A single degree-2 index, the product of two normalized differences from the red-edge bands, already reaches 96.26\% accuracy, and using eight indices only raises this to 97.70\%. In every case the chosen features are degree-2 products built from bands $b_4$ through $b_8$, which suggests that the discriminative signal comes from spectral \emph{interactions} rather than individual band ratios. Because the indices involve only simple arithmetic, they can be deployed directly in platforms like Google Earth Engine. The same approach works for other sensors and classification tasks, and an open-source implementation (\texttt{ndindex}) is available.
AutoCount: Unsupervised Segmentation and Counting of Organs in Field Images
Jul 17, 2020



Abstract:Counting plant organs such as heads or tassels from outdoor imagery is a popular benchmark computer vision task in plant phenotyping, which has been previously investigated in the literature using state-of-the-art supervised deep learning techniques. However, the annotation of organs in field images is time-consuming and prone to errors. In this paper, we propose a fully unsupervised technique for counting dense objects such as plant organs. We use a convolutional network-based unsupervised segmentation method followed by two post-hoc optimization steps. The proposed technique is shown to provide competitive counting performance on a range of organ counting tasks in sorghum (S. bicolor) and wheat (T. aestivum) with no dataset-dependent tuning or modifications.
Crop Lodging Prediction from UAV-Acquired Images of Wheat and Canola using a DCNN Augmented with Handcrafted Texture Features
Jun 18, 2019
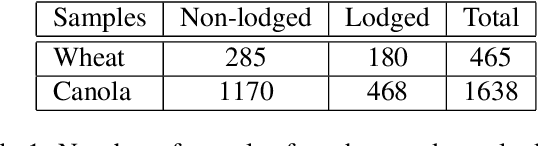
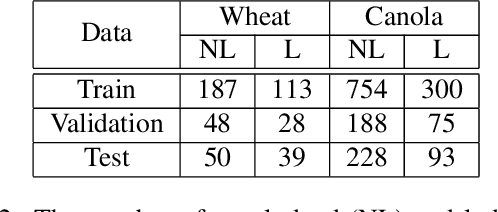

Abstract:Lodging, the permanent bending over of food crops, leads to poor plant growth and development. Consequently, lodging results in reduced crop quality, lowers crop yield, and makes harvesting difficult. Plant breeders routinely evaluate several thousand breeding lines, and therefore, automatic lodging detection and prediction is of great value aid in selection. In this paper, we propose a deep convolutional neural network (DCNN) architecture for lodging classification using five spectral channel orthomosaic images from canola and wheat breeding trials. Also, using transfer learning, we trained 10 lodging detection models using well-established deep convolutional neural network architectures. Our proposed model outperforms the state-of-the-art lodging detection methods in the literature that use only handcrafted features. In comparison to 10 DCNN lodging detection models, our proposed model achieves comparable results while having a substantially lower number of parameters. This makes the proposed model suitable for applications such as real-time classification using inexpensive hardware for high-throughput phenotyping pipelines. The GitHub repository at https://github.com/FarhadMaleki/LodgedNet contains code and models.
DeepWheat: Estimating Phenotypic Traits from Crop Images with Deep Learning
Jan 26, 2018



Abstract:In this paper, we investigate estimating emergence and biomass traits from color images and elevation maps of wheat field plots. We employ a state-of-the-art deconvolutional network for segmentation and convolutional architectures, with residual and Inception-like layers, to estimate traits via high dimensional nonlinear regression. Evaluation was performed on two different species of wheat, grown in field plots for an experimental plant breeding study. Our framework achieves satisfactory performance with mean and standard deviation of absolute difference of 1.05 and 1.40 counts for emergence and 1.45 and 2.05 for biomass estimation. Our results for counting wheat plants from field images are better than the accuracy reported for the similar, but arguably less difficult, task of counting leaves from indoor images of rosette plants. Our results for biomass estimation, even with a very small dataset, improve upon all previously proposed approaches in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge