Stéphanie Guérit
Compressive lensless endoscopy with partial speckle scanning
Apr 22, 2021



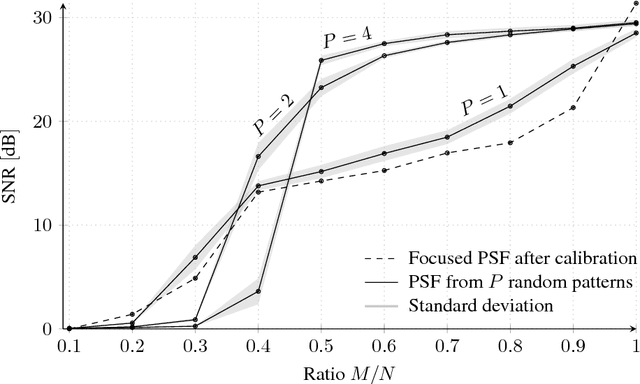
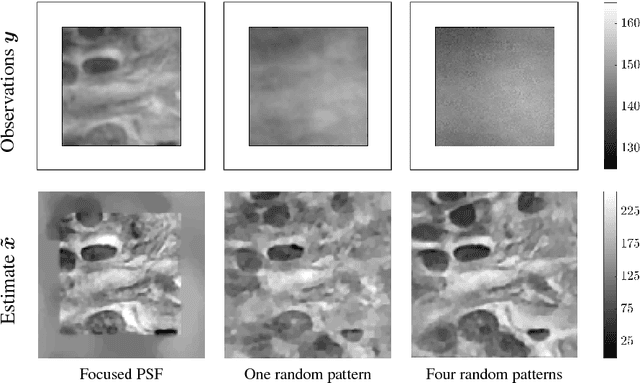
Abstract:The lensless endoscope (LE) is a promising device to acquire in vivo images at a cellular scale. The tiny size of the probe enables a deep exploration of the tissues. Lensless endoscopy with a multicore fiber (MCF) commonly uses a spatial light modulator (SLM) to coherently combine, at the output of the MCF, few hundreds of beamlets into a focus spot. This spot is subsequently scanned across the sample to generate a fluorescent image. We propose here a novel scanning scheme, partial speckle scanning (PSS), inspired by compressive sensing theory, that avoids the use of an SLM to perform fluorescent imaging in LE with reduced acquisition time. Such a strategy avoids photo-bleaching while keeping high reconstruction quality. We develop our approach on two key properties of the LE: (i) the ability to easily generate speckles, and (ii) the memory effect in MCF that allows to use fast scan mirrors to shift light patterns. First, we show that speckles are sub-exponential random fields. Despite their granular structure, an appropriate choice of the reconstruction parameters makes them good candidates to build efficient sensing matrices. Then, we numerically validate our approach and apply it on experimental data. The proposed sensing technique outperforms conventional raster scanning: higher reconstruction quality is achieved with far fewer observations. For a fixed reconstruction quality, our speckle scanning approach is faster than compressive sensing schemes which require to change the speckle pattern for each observation.
Compressive Sampling Approach for Image Acquisition with Lensless Endoscope
Oct 30, 2018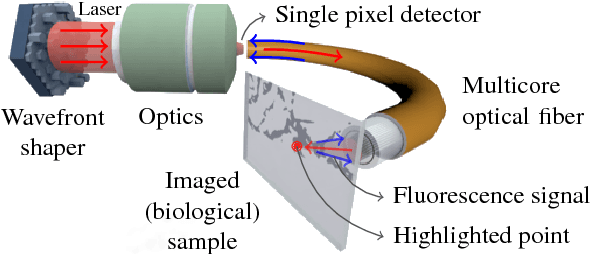
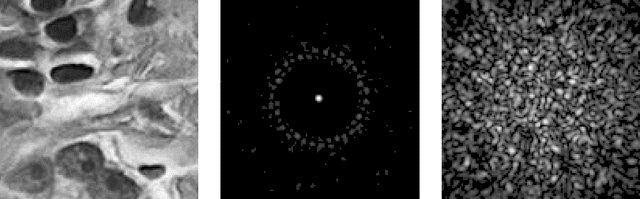
Abstract:The lensless endoscope is a promising device designed to image tissues in vivo at the cellular scale. The traditional acquisition setup consists in raster scanning during which the focused light beam from the optical fiber illuminates sequentially each pixel of the field of view (FOV). The calibration step to focus the beam and the sampling scheme both take time. In this preliminary work, we propose a scanning method based on compressive sampling theory. The method does not rely on a focused beam but rather on the random illumination patterns generated by the single-mode fibers. Experiments are performed on synthetic data for different compression rates (from 10 to 100% of the FOV).
Blind Deconvolution of PET Images using Anatomical Priors
Aug 05, 2016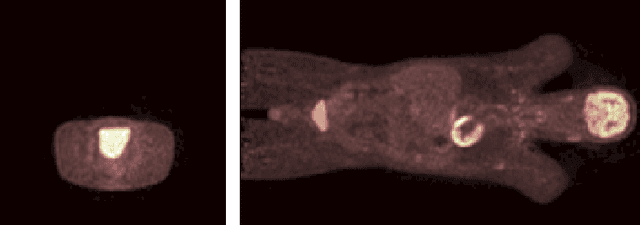
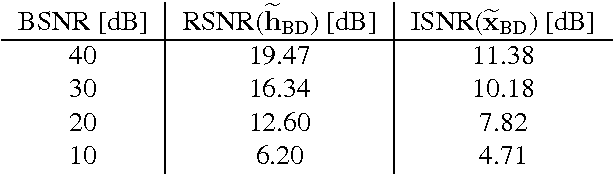
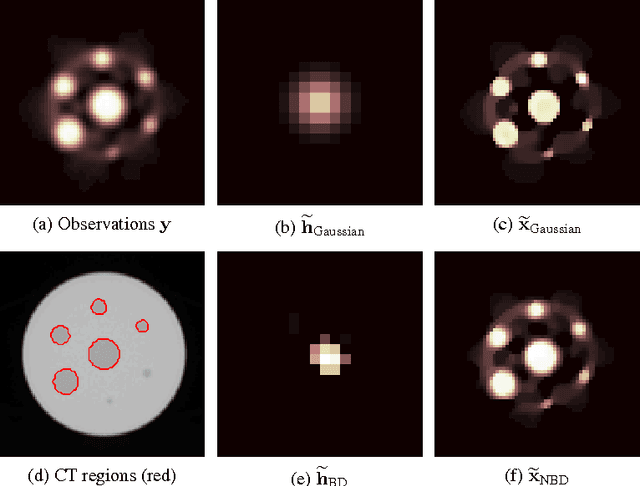
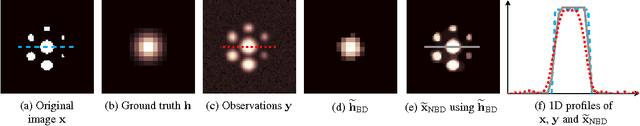
Abstract:Images from positron emission tomography (PET) provide metabolic information about the human body. They present, however, a spatial resolution that is limited by physical and instrumental factors often modeled by a blurring function. Since this function is typically unknown, blind deconvolution (BD) techniques are needed in order to produce a useful restored PET image. In this work, we propose a general BD technique that restores a low resolution blurry image using information from data acquired with a high resolution modality (e.g., CT-based delineation of regions with uniform activity in PET images). The proposed BD method is validated on synthetic and actual phantoms.
Post-Reconstruction Deconvolution of PET Images by Total Generalized Variation Regularization
Jun 16, 2015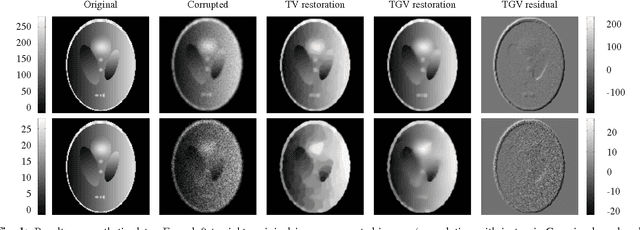
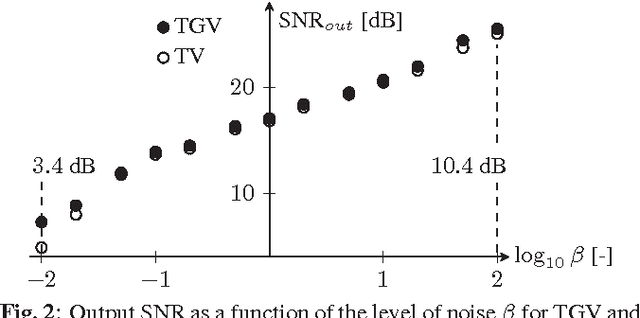
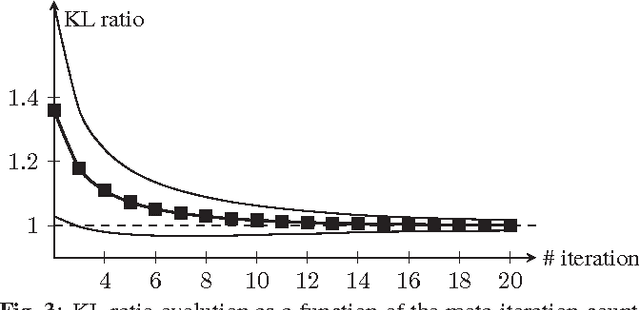
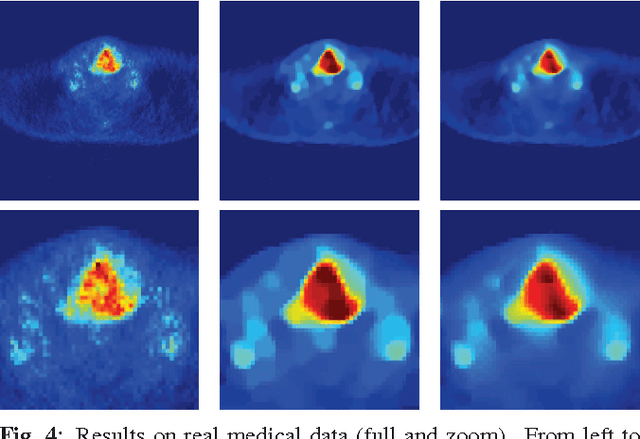
Abstract:Improving the quality of positron emission tomography (PET) images, affected by low resolution and high level of noise, is a challenging task in nuclear medicine and radiotherapy. This work proposes a restoration method, achieved after tomographic reconstruction of the images and targeting clinical situations where raw data are often not accessible. Based on inverse problem methods, our contribution introduces the recently developed total generalized variation (TGV) norm to regularize PET image deconvolution. Moreover, we stabilize this procedure with additional image constraints such as positivity and photometry invariance. A criterion for updating and adjusting automatically the regularization parameter in case of Poisson noise is also presented. Experiments are conducted on both synthetic data and real patient images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge