Shreshth Saini
Indian Institute of Technology Jodhpur
TABES: Trajectory-Aware Backward-on-Entropy Steering for Masked Diffusion Models
Jan 30, 2026Abstract:Masked Diffusion Models (MDMs) have emerged as a promising non-autoregressive paradigm for generative tasks, offering parallel decoding and bidirectional context utilization. However, current sampling methods rely on simple confidence-based heuristics that ignore the long-term impact of local decisions, leading to trajectory lock-in where early hallucinations cascade into global incoherence. While search-based methods mitigate this, they incur prohibitive computational costs ($O(K)$ forward passes per step). In this work, we propose Backward-on-Entropy (BoE) Steering, a gradient-guided inference framework that approximates infinite-horizon lookahead via a single backward pass. We formally derive the Token Influence Score (TIS) from a first-order expansion of the trajectory cost functional, proving that the gradient of future entropy with respect to input embeddings serves as an optimal control signal for minimizing uncertainty. To ensure scalability, we introduce \texttt{ActiveQueryAttention}, a sparse adjoint primitive that exploits the structure of the masking objective to reduce backward pass complexity. BoE achieves a superior Pareto frontier for inference-time scaling compared to existing unmasking methods, demonstrating that gradient-guided steering offers a mathematically principled and efficient path to robust non-autoregressive generation. We will release the code.
HIDRO-VQA: High Dynamic Range Oracle for Video Quality Assessment
Nov 18, 2023Abstract:We introduce HIDRO-VQA, a no-reference (NR) video quality assessment model designed to provide precise quality evaluations of High Dynamic Range (HDR) videos. HDR videos exhibit a broader spectrum of luminance, detail, and color than Standard Dynamic Range (SDR) videos. As HDR content becomes increasingly popular, there is a growing demand for video quality assessment (VQA) algorithms that effectively address distortions unique to HDR content. To address this challenge, we propose a self-supervised contrastive fine-tuning approach to transfer quality-aware features from the SDR to the HDR domain, utilizing unlabeled HDR videos. Our findings demonstrate that self-supervised pre-trained neural networks on SDR content can be further fine-tuned in a self-supervised setting using limited unlabeled HDR videos to achieve state-of-the-art performance on the only publicly available VQA database for HDR content, the LIVE-HDR VQA database. Moreover, our algorithm can be extended to the Full Reference VQA setting, also achieving state-of-the-art performance. Our code is available publicly at https://github.com/avinabsaha/HIDRO-VQA.
SLAe-Net: Multi-Scale Multi-Level Attention embedded Network for Retinal Vessel Segmentation
Sep 05, 2021



Abstract:Segmentation plays a crucial role in diagnosis. Studying the retinal vasculatures from fundus images help identify early signs of many crucial illnesses such as diabetic retinopathy. Due to the varying shape, size, and patterns of retinal vessels, along with artefacts and noises in fundus images, no one-stage method can accurately segment retinal vessels. In this work, we propose a multi-scale, multi-level attention embedded CNN architecture ((M)SLAe-Net) to address the issue of multi-stage processing for robust and precise segmentation of retinal vessels. We do this by extracting features at multiple scales and multiple levels of the network, enabling our model to holistically extracts the local and global features. Multi-scale features are extracted using our novel dynamic dilated pyramid pooling (D-DPP) module. We also aggregate the features from all the network levels. These effectively resolved the issues of varying shapes and artefacts and hence the need for multiple stages. To assist in better pixel-level classification, we use the Squeeze and Attention(SA) module, a smartly adapted version of the Squeeze and Excitation(SE) module for segmentation tasks in our network to facilitate pixel-group attention. Our unique network design and novel D-DPP module with efficient task-specific loss function for thin vessels enabled our model for better cross data performance. Exhaustive experimental results on DRIVE, STARE, HRF, and CHASE-DB1 show the superiority of our method.
Detector-SegMentor Network for Skin Lesion Localization and Segmentation
May 13, 2020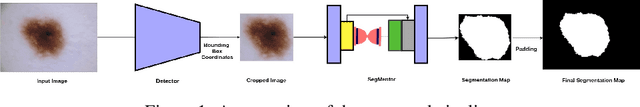
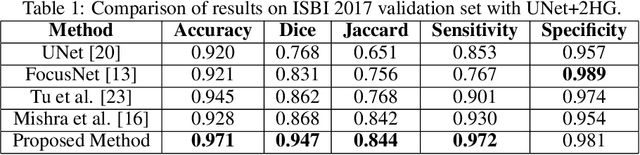
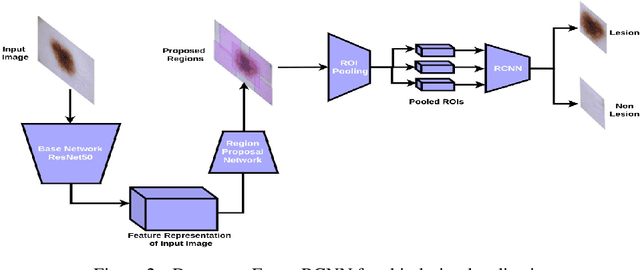
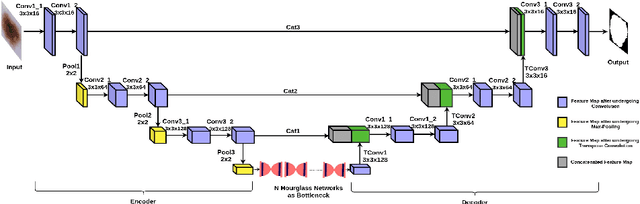
Abstract:Melanoma is a life-threatening form of skin cancer when left undiagnosed at the early stages. Although there are more cases of non-melanoma cancer than melanoma cancer, melanoma cancer is more deadly. Early detection of melanoma is crucial for the timely diagnosis of melanoma cancer and prohibit its spread to distant body parts. Segmentation of skin lesion is a crucial step in the classification of melanoma cancer from the cancerous lesions in dermoscopic images. Manual segmentation of dermoscopic skin images is very time consuming and error-prone resulting in an urgent need for an intelligent and accurate algorithm. In this study, we propose a simple yet novel network-in-network convolution neural network(CNN) based approach for segmentation of the skin lesion. A Faster Region-based CNN (Faster RCNN) is used for preprocessing to predict bounding boxes of the lesions in the whole image which are subsequently cropped and fed into the segmentation network to obtain the lesion mask. The segmentation network is a combination of the UNet and Hourglass networks. We trained and evaluated our models on ISIC 2018 dataset and also cross-validated on PH\textsuperscript{2} and ISBI 2017 datasets. Our proposed method surpassed the state-of-the-art with Dice Similarity Coefficient of 0.915 and Accuracy 0.959 on ISIC 2018 dataset and Dice Similarity Coefficient of 0.947 and Accuracy 0.971 on ISBI 2017 dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge