Shen-Chieh Tai
A Closer Look at Spatial-Slice Features Learning for COVID-19 Detection
Apr 02, 2024



Abstract:Conventional Computed Tomography (CT) imaging recognition faces two significant challenges: (1) There is often considerable variability in the resolution and size of each CT scan, necessitating strict requirements for the input size and adaptability of models. (2) CT-scan contains large number of out-of-distribution (OOD) slices. The crucial features may only be present in specific spatial regions and slices of the entire CT scan. How can we effectively figure out where these are located? To deal with this, we introduce an enhanced Spatial-Slice Feature Learning (SSFL++) framework specifically designed for CT scan. It aim to filter out a OOD data within whole CT scan, enabling our to select crucial spatial-slice for analysis by reducing 70% redundancy totally. Meanwhile, we proposed Kernel-Density-based slice Sampling (KDS) method to improve the stability when training and inference stage, therefore speeding up the rate of convergence and boosting performance. As a result, the experiments demonstrate the promising performance of our model using a simple EfficientNet-2D (E2D) model, even with only 1% of the training data. The efficacy of our approach has been validated on the COVID-19-CT-DB datasets provided by the DEF-AI-MIA workshop, in conjunction with CVPR 2024. Our source code will be made available.
Simple 2D Convolutional Neural Network-based Approach for COVID-19 Detection
Mar 17, 2024



Abstract:This study explores the use of deep learning techniques for analyzing lung Computed Tomography (CT) images. Classic deep learning approaches face challenges with varying slice counts and resolutions in CT images, a diversity arising from the utilization of assorted scanning equipment. Typically, predictions are made on single slices which are then combined for a comprehensive outcome. Yet, this method does not incorporate learning features specific to each slice, leading to a compromise in effectiveness. To address these challenges, we propose an advanced Spatial-Slice Feature Learning (SSFL++) framework specifically tailored for CT scans. It aims to filter out out-of-distribution (OOD) data within the entire CT scan, allowing us to select essential spatial-slice features for analysis by reducing data redundancy by 70\%. Additionally, we introduce a Kernel-Density-based slice Sampling (KDS) method to enhance stability during training and inference phases, thereby accelerating convergence and enhancing overall performance. Remarkably, our experiments reveal that our model achieves promising results with a simple EfficientNet-2D (E2D) model. The effectiveness of our approach is confirmed on the COVID-19-CT-DB datasets provided by the DEF-AI-MIA workshop.
Spatiotemporal Feature Learning Based on Two-Step LSTM and Transformer for CT Scans
Jul 08, 2022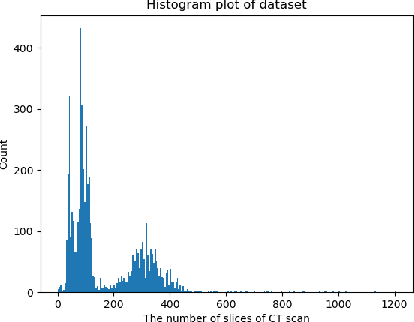
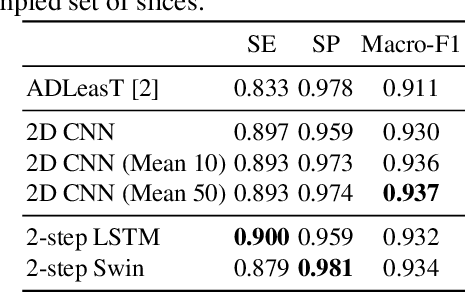
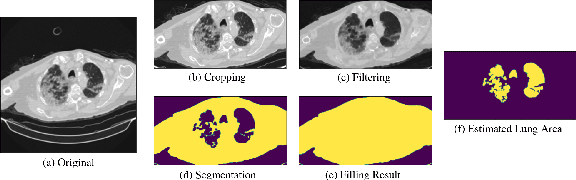
Abstract:Computed tomography (CT) imaging could be very practical for diagnosing various diseases. However, the nature of the CT images is even more diverse since the resolution and number of the slices of a CT scan are determined by the machine and its settings. Conventional deep learning models are hard to tickle such diverse data since the essential requirement of the deep neural network is the consistent shape of the input data. In this paper, we propose a novel, effective, two-step-wise approach to tickle this issue for COVID-19 symptom classification thoroughly. First, the semantic feature embedding of each slice for a CT scan is extracted by conventional backbone networks. Then, we proposed a long short-term memory (LSTM) and Transformer-based sub-network to deal with temporal feature learning, leading to spatiotemporal feature representation learning. In this fashion, the proposed two-step LSTM model could prevent overfitting, as well as increase performance. Comprehensive experiments reveal that the proposed two-step method not only shows excellent performance but also could be compensated for each other. More specifically, the two-step LSTM model has a lower false-negative rate, while the 2-step Swin model has a lower false-positive rate. In summary, it is suggested that the model ensemble could be adopted for more stable and promising performance in real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge