Shangpeng Sun
Predictive Pattern Recognition Techniques Towards Spatiotemporal Representation of Plant Growth in Simulated and Controlled Environments: A Comprehensive Review
Dec 13, 2024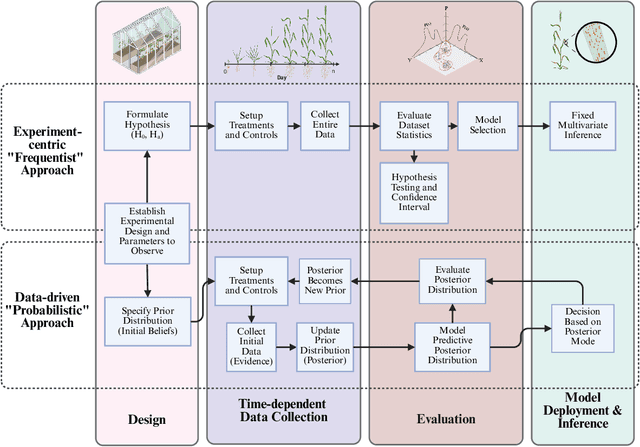
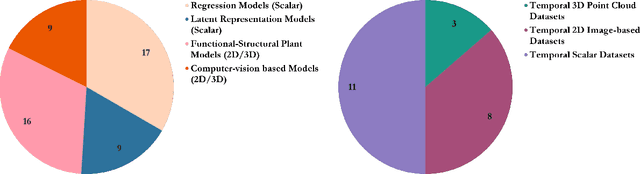
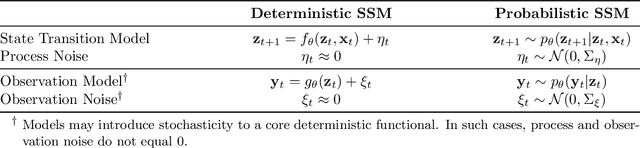
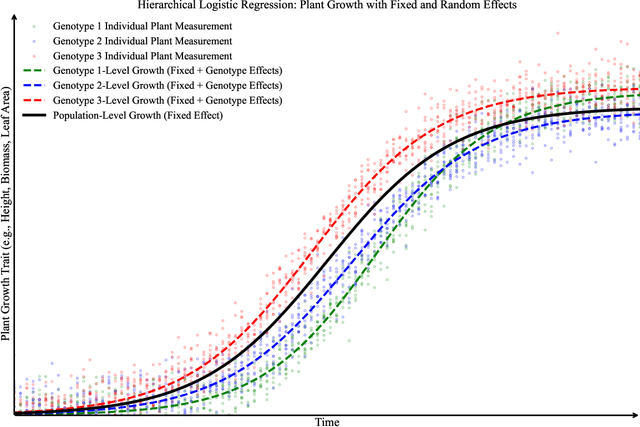
Abstract:Accurate predictions and representations of plant growth patterns in simulated and controlled environments are important for addressing various challenges in plant phenomics research. This review explores various works on state-of-the-art predictive pattern recognition techniques, focusing on the spatiotemporal modeling of plant traits and the integration of dynamic environmental interactions. We provide a comprehensive examination of deterministic, probabilistic, and generative modeling approaches, emphasizing their applications in high-throughput phenotyping and simulation-based plant growth forecasting. Key topics include regressions and neural network-based representation models for the task of forecasting, limitations of existing experiment-based deterministic approaches, and the need for dynamic frameworks that incorporate uncertainty and evolving environmental feedback. This review surveys advances in 2D and 3D structured data representations through functional-structural plant models and conditional generative models. We offer a perspective on opportunities for future works, emphasizing the integration of domain-specific knowledge to data-driven methods, improvements to available datasets, and the implementation of these techniques toward real-world applications.
Generative Plant Growth Simulation from Sequence-Informed Environmental Conditions
May 23, 2024Abstract:A plant growth simulation can be characterized as a reconstructed visual representation of a plant or plant system. The phenotypic characteristics and plant structures are controlled by the scene environment and other contextual attributes. Considering the temporal dependencies and compounding effects of various factors on growth trajectories, we formulate a probabilistic approach to the simulation task by solving a frame synthesis and pattern recognition problem. We introduce a Sequence-Informed Plant Growth Simulation framework (SI-PGS) that employs a conditional generative model to implicitly learn a distribution of possible plant representations within a dynamic scene from a fusion of low dimensional temporal sensor and context data. Methods such as controlled latent sampling and recurrent output connections are used to improve coherence in plant structures between frames of predictions. In this work, we demonstrate that SI-PGS is able to capture temporal dependencies and continuously generate realistic frames of a plant scene.
Eff-3DPSeg: 3D organ-level plant shoot segmentation using annotation-efficient point clouds
Dec 20, 2022



Abstract:Reliable and automated 3D plant shoot segmentation is a core prerequisite for the extraction of plant phenotypic traits at the organ level. Combining deep learning and point clouds can provide effective ways to address the challenge. However, fully supervised deep learning methods require datasets to be point-wise annotated, which is extremely expensive and time-consuming. In our work, we proposed a novel weakly supervised framework, Eff-3DPSeg, for 3D plant shoot segmentation. First, high-resolution point clouds of soybean were reconstructed using a low-cost photogrammetry system, and the Meshlab-based Plant Annotator was developed for plant point cloud annotation. Second, a weakly-supervised deep learning method was proposed for plant organ segmentation. The method contained: (1) Pretraining a self-supervised network using Viewpoint Bottleneck loss to learn meaningful intrinsic structure representation from the raw point clouds; (2) Fine-tuning the pre-trained model with about only 0.5% points being annotated to implement plant organ segmentation. After, three phenotypic traits (stem diameter, leaf width, and leaf length) were extracted. To test the generality of the proposed method, the public dataset Pheno4D was included in this study. Experimental results showed that the weakly-supervised network obtained similar segmentation performance compared with the fully-supervised setting. Our method achieved 95.1%, 96.6%, 95.8% and 92.2% in the Precision, Recall, F1-score, and mIoU for stem leaf segmentation and 53%, 62.8% and 70.3% in the AP, AP@25, and AP@50 for leaf instance segmentation. This study provides an effective way for characterizing 3D plant architecture, which will become useful for plant breeders to enhance selection processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge