Shams Mehdi
Enhanced Sampling with Machine Learning: A Review
Jun 16, 2023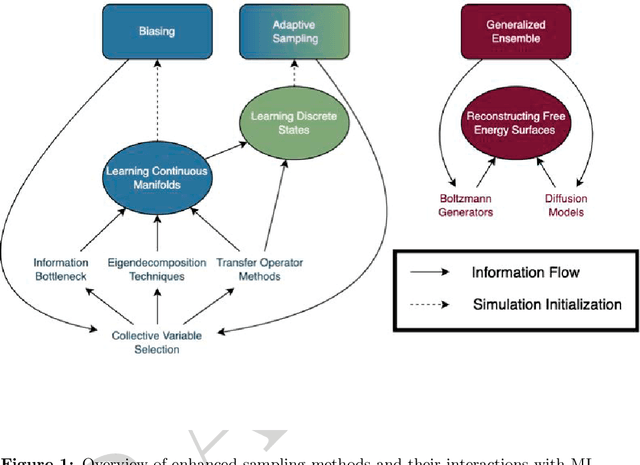



Abstract:Molecular dynamics (MD) enables the study of physical systems with excellent spatiotemporal resolution but suffers from severe time-scale limitations. To address this, enhanced sampling methods have been developed to improve exploration of configurational space. However, implementing these is challenging and requires domain expertise. In recent years, integration of machine learning (ML) techniques in different domains has shown promise, prompting their adoption in enhanced sampling as well. Although ML is often employed in various fields primarily due to its data-driven nature, its integration with enhanced sampling is more natural with many common underlying synergies. This review explores the merging of ML and enhanced MD by presenting different shared viewpoints. It offers a comprehensive overview of this rapidly evolving field, which can be difficult to stay updated on. We highlight successful strategies like dimensionality reduction, reinforcement learning, and flow-based methods. Finally, we discuss open problems at the exciting ML-enhanced MD interface.
Thermodynamics of Interpretation
Jun 27, 2022



Abstract:Over the past few years, different types of data-driven Artificial Intelligence (AI) techniques have been widely adopted in various domains of science for generating predictive black-box models. However, because of their black-box nature, it is crucial to establish trust in these models before accepting them as accurate. One way of achieving this goal is through the implementation of a post-hoc interpretation scheme that can put forward the reasons behind a black-box model prediction. In this work, we propose a classical thermodynamics inspired approach for this purpose: Thermodynamically Explainable Representations of AI and other black-box Paradigms (TERP). TERP works by constructing a linear, local surrogate model that approximates the behaviour of the black-box model within a small neighborhood around the instance being explained. By employing a simple forward feature selection Monte Carlo algorithm, TERP assigns an interpretability free energy score to all the possible surrogate models in order to choose an optimal interpretation. Additionally, we validate TERP as a generally applicable method by successfully interpreting four different classes of black-box models trained on datasets coming from relevant domains, including classifying images, predicting heart disease and classifying biomolecular conformations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge