Seyedjafar Shojaerazavi
A novel unsupervised covid lung lesion segmentation based on the lung tissue identification
Feb 24, 2022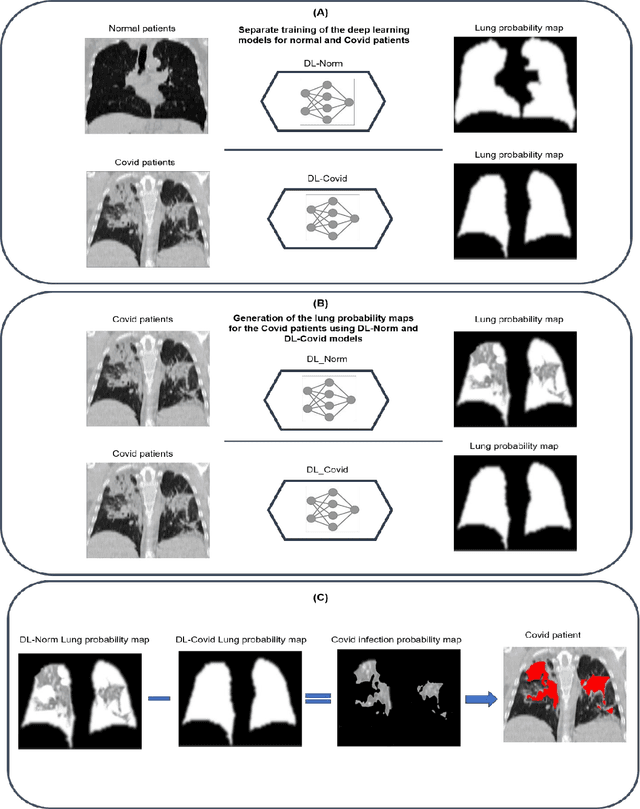
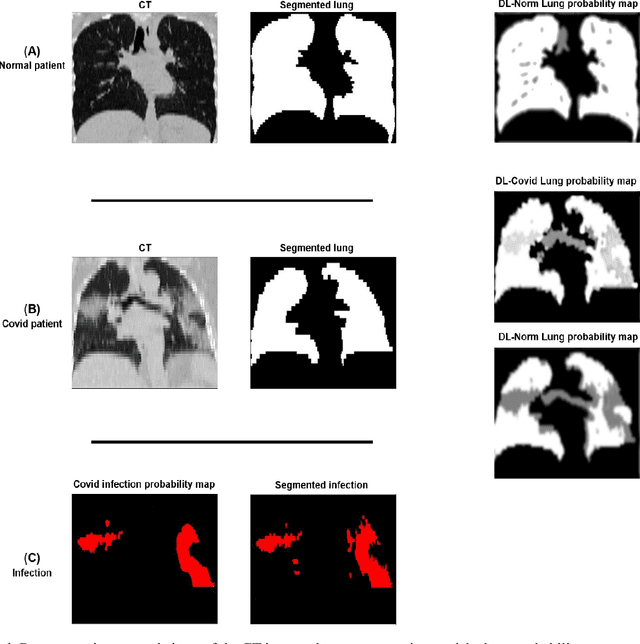

Abstract:This study aimed to evaluate the performance of a novel unsupervised deep learning-based framework for automated infections lesion segmentation from CT images of Covid patients. In the first step, two residual networks were independently trained to identify the lung tissue for normal and Covid patients in a supervised manner. These two models, referred to as DL-Covid and DL-Norm for Covid-19 and normal patients, respectively, generate the voxel-wise probability maps for lung tissue identification. To detect Covid lesions, the CT image of the Covid patient is processed by the DL-Covid and DL-Norm models to obtain two lung probability maps. Since the DL-Norm model is not familiar with Covid infections within the lung, this model would assign lower probabilities to the lesions than the DL-Covid. Hence, the probability maps of the Covid infections could be generated through the subtraction of the two lung probability maps obtained from the DL-Covid and DL-Norm models. Manual lesion segmentation of 50 Covid-19 CT images was used to assess the accuracy of the unsupervised lesion segmentation approach. The Dice coefficients of 0.985 and 0.978 were achieved for the lung segmentation of normal and Covid patients in the external validation dataset, respectively. Quantitative results of infection segmentation by the proposed unsupervised method showed the Dice coefficient and Jaccard index of 0.67 and 0.60, respectively. Quantitative evaluation of the proposed unsupervised approach for Covid-19 infectious lesion segmentation showed relatively satisfactory results. Since this framework does not require any annotated dataset, it could be used to generate very large training samples for the supervised machine learning algorithms dedicated to noisy and/or weakly annotated datasets.
Automated lung segmentation from CT images of normal and COVID-19 pneumonia patients
Apr 05, 2021
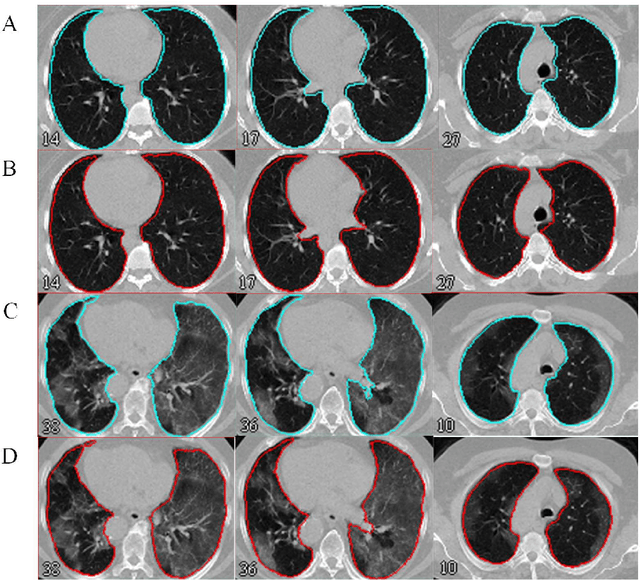
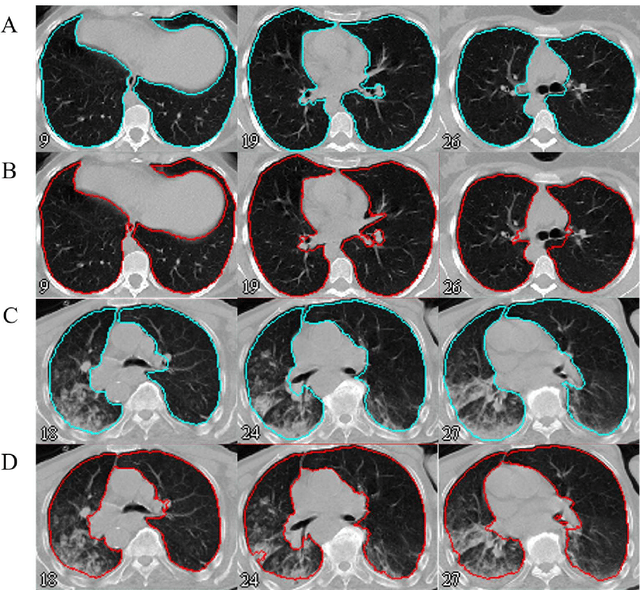
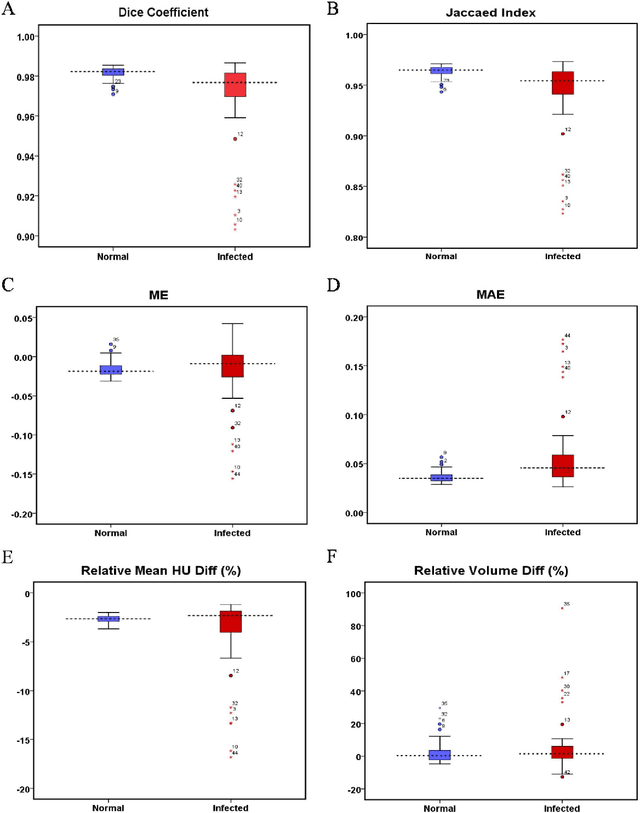
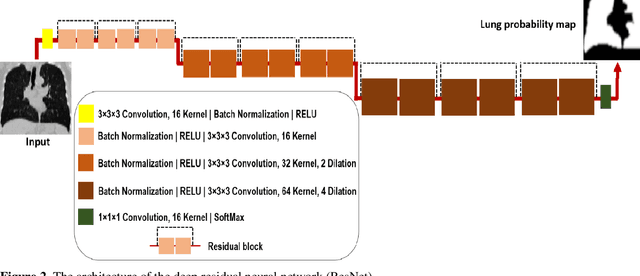
Abstract:Automated semantic image segmentation is an essential step in quantitative image analysis and disease diagnosis. This study investigates the performance of a deep learning-based model for lung segmentation from CT images for normal and COVID-19 patients. Chest CT images and corresponding lung masks of 1200 confirmed COVID-19 cases were used for training a residual neural network. The reference lung masks were generated through semi-automated/manual segmentation of the CT images. The performance of the model was evaluated on two distinct external test datasets including 120 normal and COVID-19 subjects, and the results of these groups were compared to each other. Different evaluation metrics such as dice coefficient (DSC), mean absolute error (MAE), relative mean HU difference, and relative volume difference were calculated to assess the accuracy of the predicted lung masks. The proposed deep learning method achieved DSC of 0.980 and 0.971 for normal and COVID-19 subjects, respectively, demonstrating significant overlap between predicted and reference lung masks. Moreover, MAEs of 0.037 HU and 0.061 HU, relative mean HU difference of -2.679% and -4.403%, and relative volume difference of 2.405% and 5.928% were obtained for normal and COVID-19 subjects, respectively. The comparable performance in lung segmentation of the normal and COVID-19 patients indicates the accuracy of the model for the identification of the lung tissue in the presence of the COVID-19 induced infections (though slightly better performance was observed for normal patients). The promising results achieved by the proposed deep learning-based model demonstrated its reliability in COVID-19 lung segmentation. This prerequisite step would lead to a more efficient and robust pneumonia lesion analysis.
Quantitative analysis of image quality in low-dose CT imaging for Covid-19 patients
Feb 16, 2021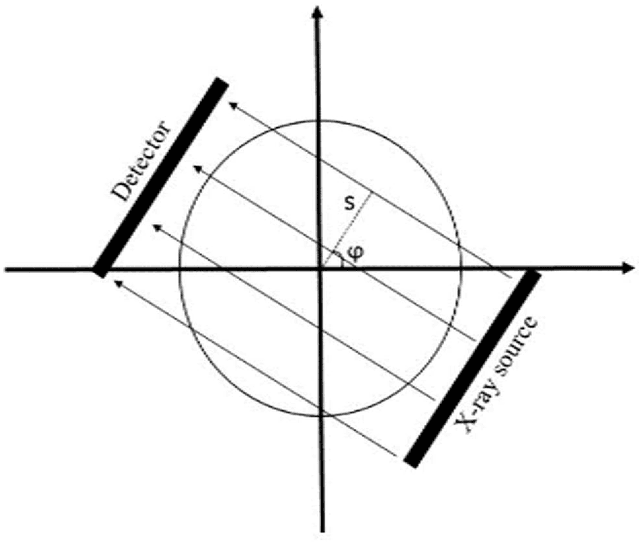
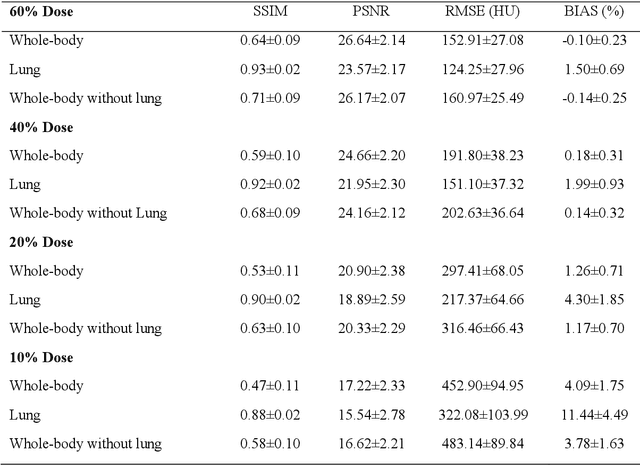
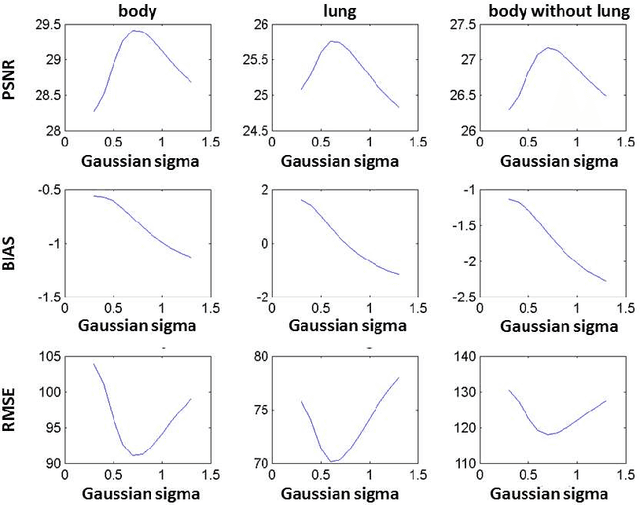
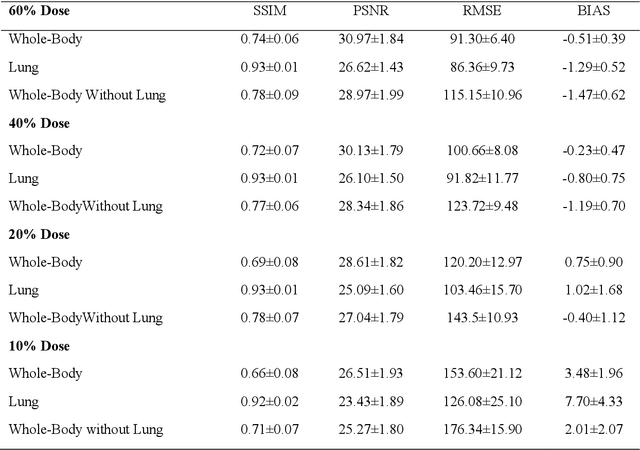
Abstract:We set out to simulate four reduced dose-levels (60%-dose, 40%-dose, 20%-dose, and 10%-dose) of standard CT imaging using Beer-Lambert's law across 49 patients infected with COVID-19. Then, three denoising filters, namely Gaussian, Bilateral, and Median, were applied to the different low-dose CT images, the quality of which was assessed prior to and after the application of the various filters via calculation of peak signal-to-noise ratio (PSNR), root mean square error (RMSE), structural similarity index measure (SSIM), and relative CT-value bias, separately for the lung tissue and whole-body. The quantitative evaluation indicated that 10%-dose CT images have inferior quality (with RMSE=322.1-+104.0 HU and bias=11.44-+4.49% in the lung) even after the application of the denoising filters. The bilateral filter exhibited superior performance to suppress the noise and recover the underlying signals in low-dose CT images compared to the other denoising techniques. The bilateral filter led to RMSE and bias of 100.21-+16.47 HU, -0.21-+1.20%, respectively in the lung regions for 20%-dose CT images compared to the Gaussian filter with RMSE=103.46-+15.70 HU and bias=1.02-+1.68%, median filter with RMSE=129.60-+18.09 HU and bias=-6.15-+2.24%, and the nonfiltered 20%-dose CT with RMSE=217.37-+64.66 HU and bias=4.30-+1.85%. In conclusion, the 20%-dose CT imaging followed by the bilateral filtering introduced a reasonable compromise between image quality and patient dose reduction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge