Sebastian Magierowski
LRDB: LSTM Raw data DNA Base-caller based on long-short term models in an active learning environment
Mar 15, 2023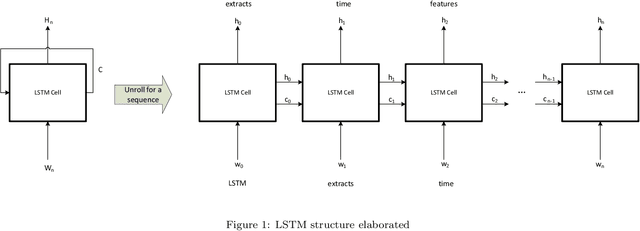
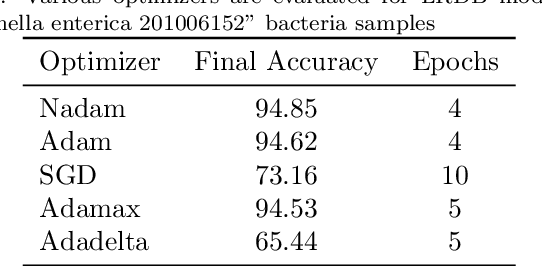
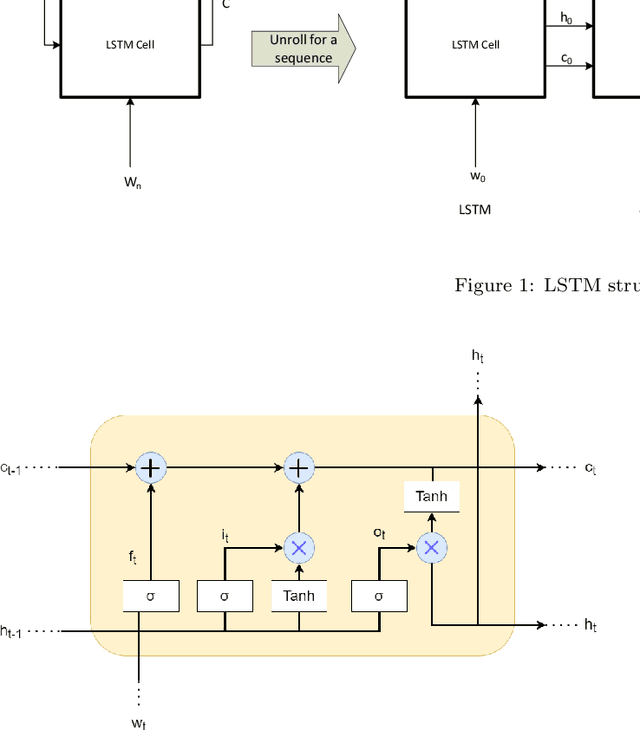
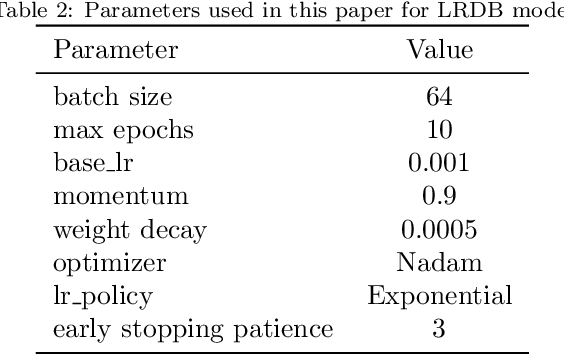
Abstract:The first important step in extracting DNA characters is using the output data of MinION devices in the form of electrical current signals. Various cutting-edge base callers use this data to detect the DNA characters based on the input. In this paper, we discuss several shortcomings of prior base callers in the case of time-critical applications, privacy-aware design, and the problem of catastrophic forgetting. Next, we propose the LRDB model, a lightweight open-source model for private developments with a better read-identity (0.35% increase) for the target bacterial samples in the paper. We have limited the extent of training data and benefited from the transfer learning algorithm to make the active usage of the LRDB viable in critical applications. Henceforth, less training time for adapting to new DNA samples (in our case, Bacterial samples) is needed. Furthermore, LRDB can be modified concerning the user constraints as the results show a negligible accuracy loss in case of using fewer parameters. We have also assessed the noise-tolerance property, which offers about a 1.439% decline in accuracy for a 15dB noise injection, and the performance metrics show that the model executes in a medium speed range compared with current cutting-edge models.
Nanopore-Based DNA Sequencing Sensors and CMOS Readout Approaches
Oct 05, 2021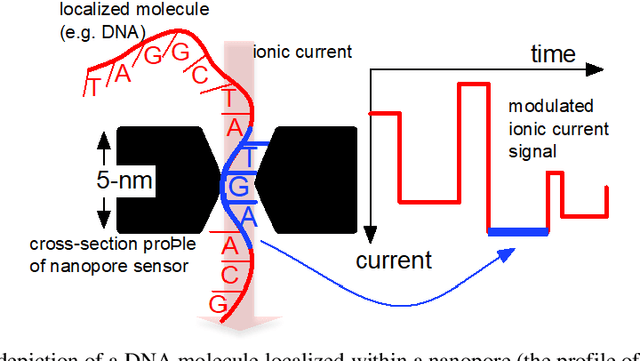
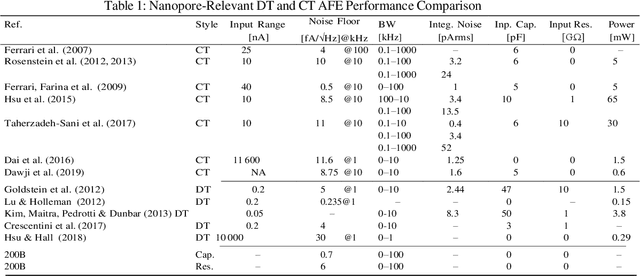
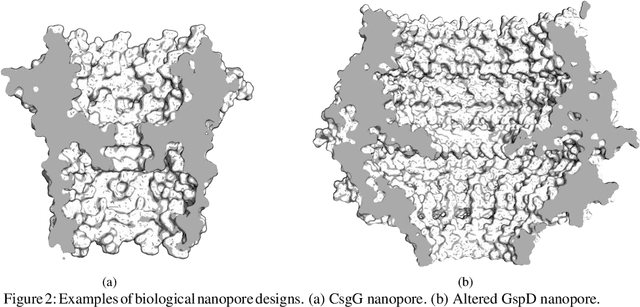
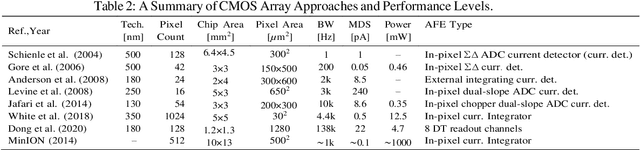
Abstract:Purpose Nanopore-based molecular sensing and measurement, specifically Deoxyribonucleic acid (DNA) sequencing, is advancing at a fast pace. Some embodiments have matured from coarse particle counters to enabling full human genome assembly. This evolution has been powered not only by improvements in the sensors themselves, but also in the assisting microelectronic Complementary Metal Oxide Semiconductor (CMOS) readout circuitry closely interfaced to them. In this light, this paper reviews established and emerging nanopore-based sensing modalities considered for DNA sequencing and CMOS microelectronic methods currently being used. Design/methodology/approach Readout and amplifier circuits which are potentially appropriate for conditioning and conversion of nanopore signals for downstream processing are studied. Furthermore, arrayed CMOS readout implementations are focused on and the relevant status of the nanopore sensor technology is reviewed as well. Findings Ion channel nanopore devices have properties unique compared with other electrochemical cells. Currently biological nanopores are the only variants reported which can be used for actual DNA sequencing. The translocation rate of DNA through such pores, the current range at which these cells operate on and the cell capacitance effect, all impose the necessity of using low noise circuits in the process of signal detection. The requirement of using in-pixel low noise circuits in turn tends to impose challenges in the implementation of large size arrays. Originality/value The study presents an overview on the readout circuits used for signal acquisition in electrochemical cell arrays and investigates the specific requirements necessary for implementation of nanopore type electrochemical cell amplifiers and their associated readout electronics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge