Scott Christley
Facilitating automated conversion of scientific knowledge into scientific simulation models with the Machine Assisted Generation, Calibration, and Comparison (MAGCC) Framework
Apr 21, 2022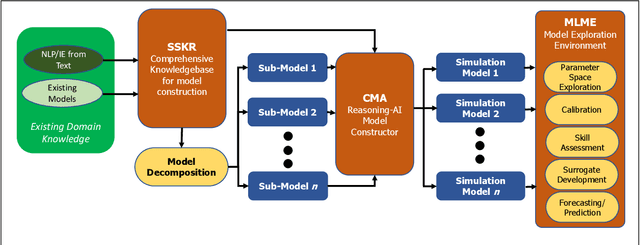
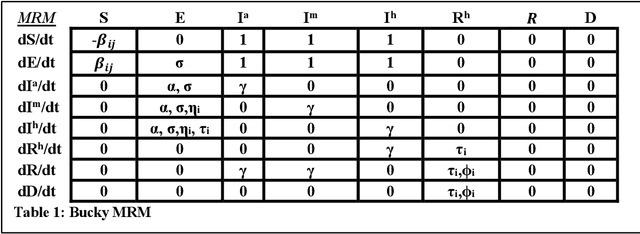
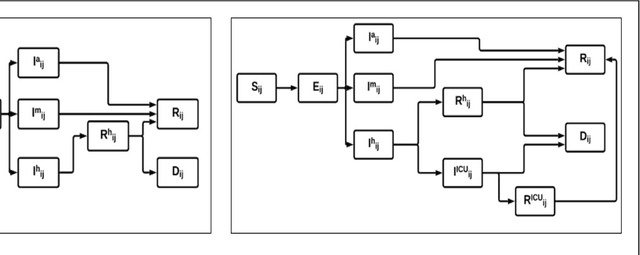
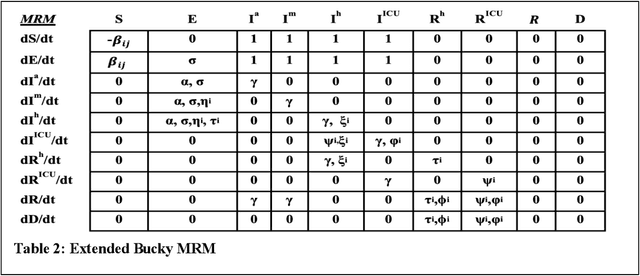
Abstract:The Machine Assisted Generation, Comparison, and Calibration (MAGCC) framework provides machine assistance and automation of recurrent crucial steps and processes in the development, implementation, testing, and use of scientific simulation models. MAGCC bridges systems for knowledge extraction via natural language processing or extracted from existing mathematical models and provides a comprehensive workflow encompassing the composition of scientific models and artificial intelligence (AI) assisted code generation. MAGCC accomplishes this through: 1) the development of a comprehensively expressive formal knowledge representation knowledgebase, the Structured Scientific Knowledge Representation (SSKR) that encompasses all the types of information needed to make any simulation model, 2) the use of an artificially intelligent logic reasoning system, the Computational Modeling Assistant (CMA), that takes information from the SSKR and generates, in a traceable fashion, model specifications across a range of simulation modeling methods, and 3) the use of the CMA to generate executable code for a simulation model from those model specifications. The MAGCC framework can be customized any scientific domain, and future work will integrate newly developed code-generating AI systems.
Dynamic Kernel Matching for Non-conforming Data: A Case Study of T-cell Receptor Datasets
Mar 18, 2021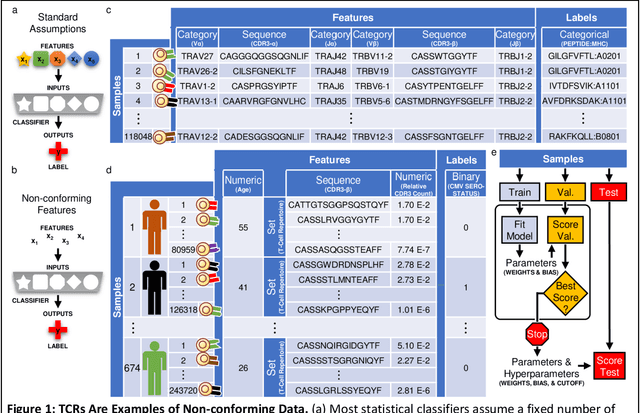
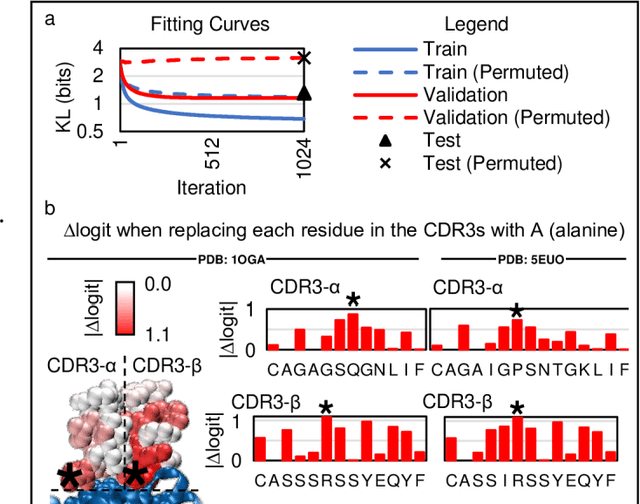
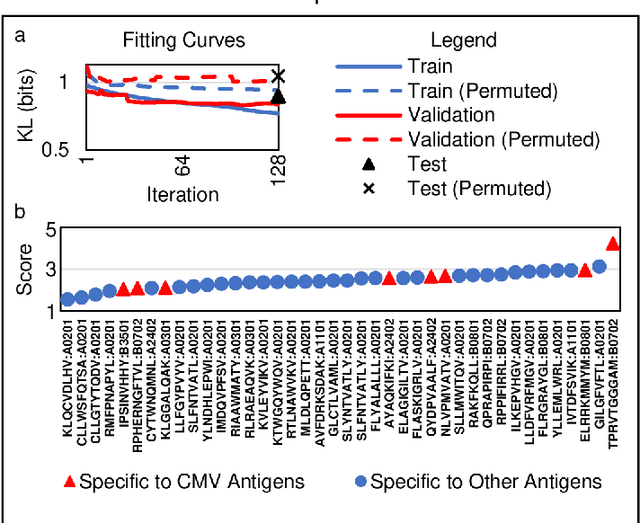
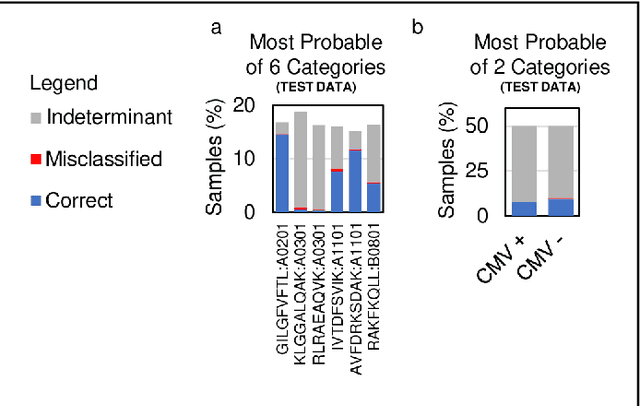
Abstract:Most statistical classifiers are designed to find patterns in data where numbers fit into rows and columns, like in a spreadsheet, but many kinds of data do not conform to this structure. To uncover patterns in non-conforming data, we describe an approach for modifying established statistical classifiers to handle non-conforming data, which we call dynamic kernel matching (DKM). As examples of non-conforming data, we consider (i) a dataset of T-cell receptor (TCR) sequences labelled by disease antigen and (ii) a dataset of sequenced TCR repertoires labelled by patient cytomegalovirus (CMV) serostatus, anticipating that both datasets contain signatures for diagnosing disease. We successfully fit statistical classifiers augmented with DKM to both datasets and report the performance on holdout data using standard metrics and metrics allowing for indeterminant diagnoses. Finally, we identify the patterns used by our statistical classifiers to generate predictions and show that these patterns agree with observations from experimental studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge