Sarwar Khan
CapST: An Enhanced and Lightweight Method for Deepfake Video Classification
Nov 07, 2023Abstract:The proliferation of deepfake videos, synthetic media produced through advanced Artificial Intelligence techniques has raised significant concerns across various sectors, encompassing realms such as politics, entertainment, and security. In response, this research introduces an innovative and streamlined model designed to classify deepfake videos generated by five distinct encoders adeptly. Our approach not only achieves state of the art performance but also optimizes computational resources. At its core, our solution employs part of a VGG19bn as a backbone to efficiently extract features, a strategy proven effective in image-related tasks. We integrate a Capsule Network coupled with a Spatial Temporal attention mechanism to bolster the model's classification capabilities while conserving resources. This combination captures intricate hierarchies among features, facilitating robust identification of deepfake attributes. Delving into the intricacies of our innovation, we introduce an existing video level fusion technique that artfully capitalizes on temporal attention mechanisms. This mechanism serves to handle concatenated feature vectors, capitalizing on the intrinsic temporal dependencies embedded within deepfake videos. By aggregating insights across frames, our model gains a holistic comprehension of video content, resulting in more precise predictions. Experimental results on an extensive benchmark dataset of deepfake videos called DFDM showcase the efficacy of our proposed method. Notably, our approach achieves up to a 4 percent improvement in accurately categorizing deepfake videos compared to baseline models, all while demanding fewer computational resources.
Detection of Diabetic Anomalies in Retinal Images using Morphological Cascading Decision Tree
Jan 07, 2020
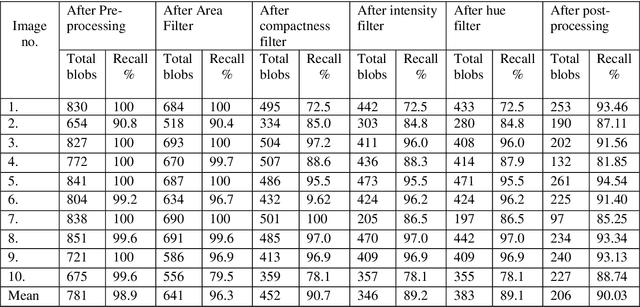
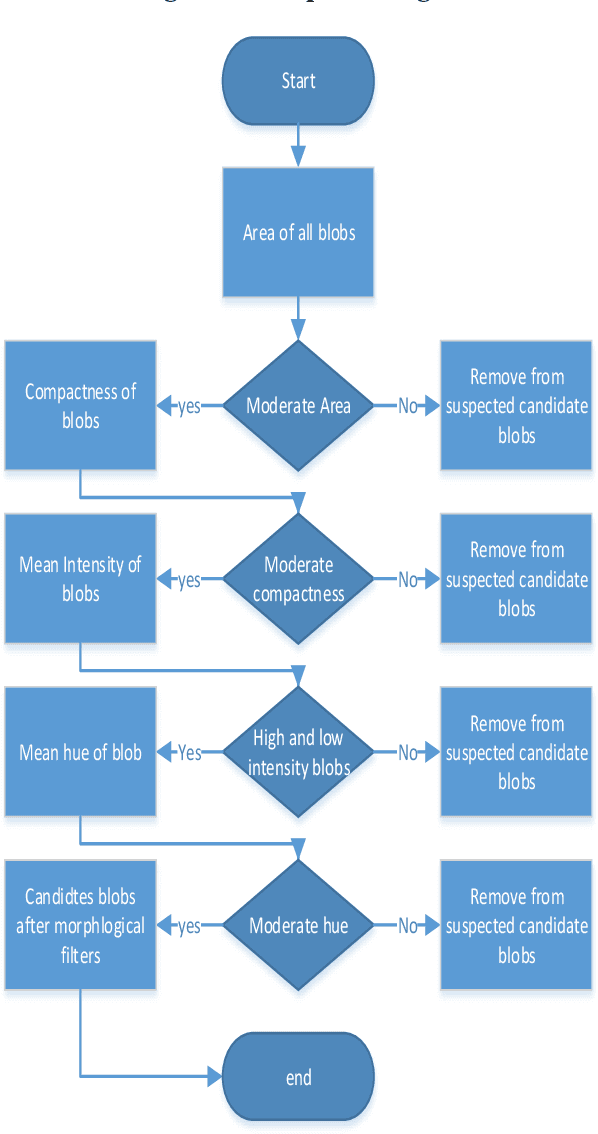
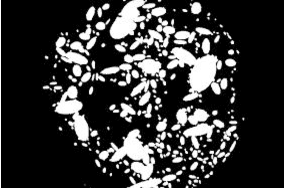
Abstract:This research aims to develop an efficient system for screening of diabetic retinopathy. Diabetic retinopathy is the major cause of blindness. Severity of diabetic retinopathy is recognized by some features, such as blood vessel area, exudates, haemorrhages and microaneurysms. To grade the disease the screening system must efficiently detect these features. In this paper we are proposing a simple and fast method for detection of diabetic retinopathy. We do pre-processing of grey-scale image and find all labelled connected components (blobs) in an image regardless of whether it is haemorrhages, exudates, vessels, optic disc or anything else. Then we apply some constraints such as compactness, area of blob, intensity and contrast for screening of candidate connectedcomponent responsible for diabetic retinopathy. We obtain our final results by doing some post processing. The results are compared with ground truths. Performance is measured by finding the recall (sensitivity). We took 10 images of dimension 500 * 752. The mean recall is 90.03%.
Macromolecule Classification Based on the Amino-acid Sequence
Jan 06, 2020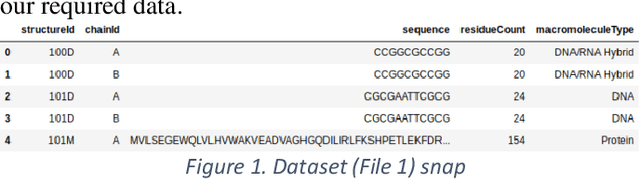
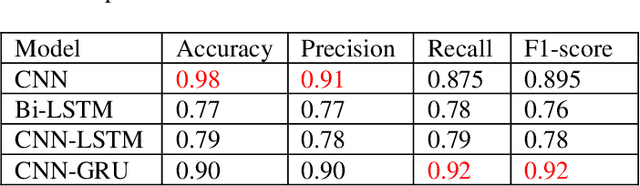
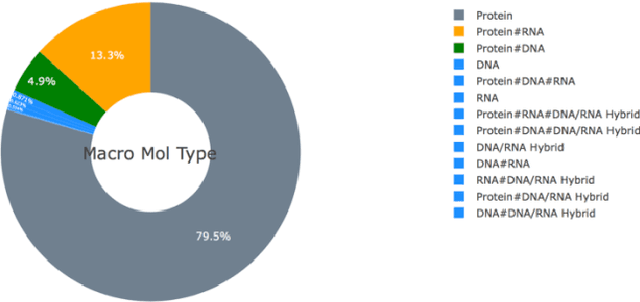
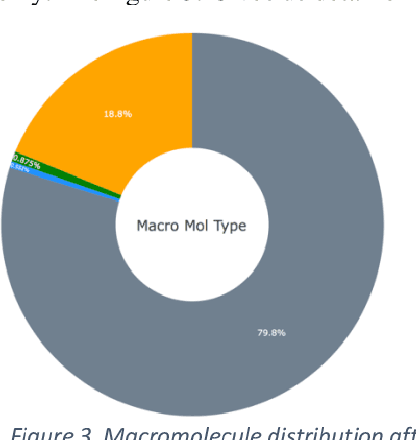
Abstract:Deep learning is playing a vital role in every field which involves data. It has emerged as a strong and efficient framework that can be applied to a broad spectrum of complex learning problems which were difficult to solve using traditional machine learning techniques in the past. In this study we focused on classification of protein sequences with deep learning techniques. The study of amino acid sequence is vital in life sciences. We used different word embedding techniques from Natural Language processing to represent the amino acid sequence as vectors. Our main goal was to classify sequences to four group of classes, that are DNA, RNA, Protein and hybrid. After several tests we have achieved almost 99% of train and test accuracy. We have experimented on CNN, LSTM, Bidirectional LSTM, and GRU.
DeepAcid: Classification of macromolecule type based on sequences of amino acids
Jul 01, 2019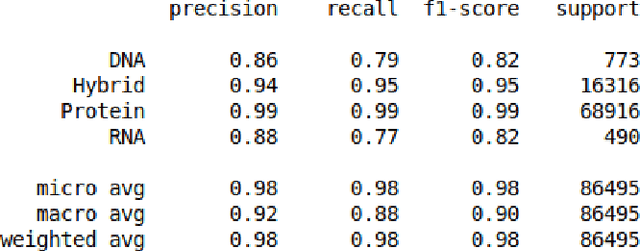
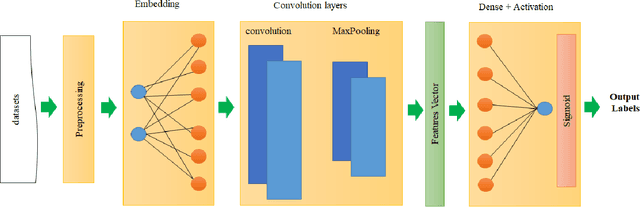
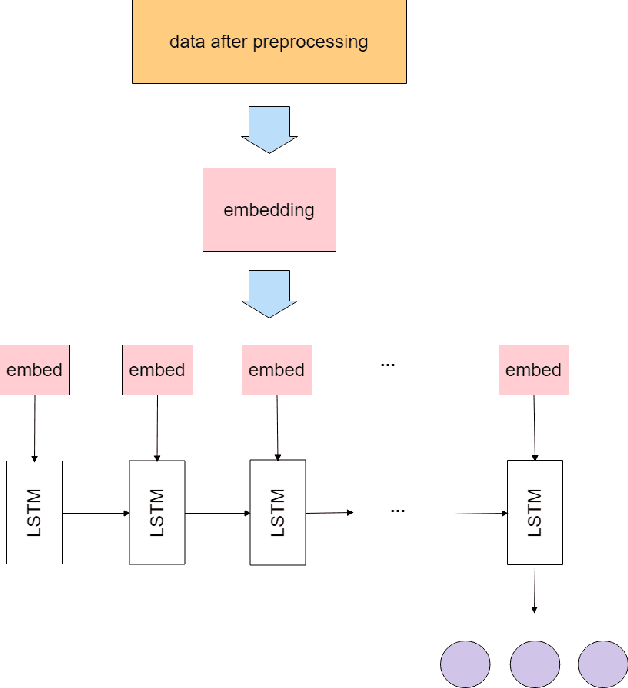
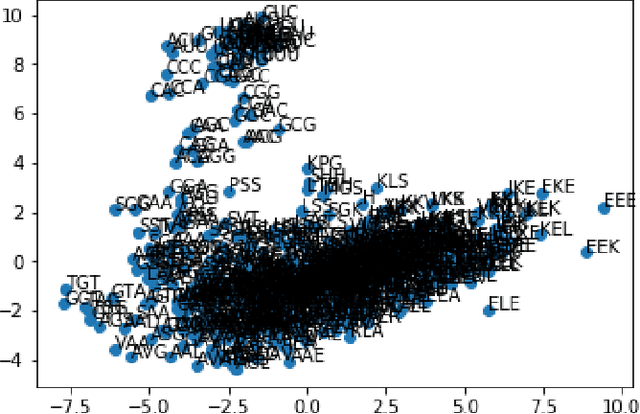
Abstract:The study of the amino acid sequence is vital in life sciences. In this paper, we are using deep learning to solve macromolecule classification problem using amino acids. Deep learning has emerged as a strong and efficient framework that can be applied to a broad spectrum of complex learning problems which were difficult to solve using traditional machine learning techniques in the past. We are using word embedding from NLP to represent the amino acid sequence as vectors. We are using different deep learning model for classification of macromolecules like CNN, LSTM, and GRU. Convolution neural network can extract features from amino acid sequences which are represented by vectors. The extracted features will be feed to a different type of model to train a robust classifier. our results show that Word2vec as embedding combine with VGG-16 has better performance than LSTM and GRU. our approach gets an error rate of 1.5%. Code is available at https://github.com/say2sarwar/DeepAcid
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge