DeepAcid: Classification of macromolecule type based on sequences of amino acids
Paper and Code
Jul 01, 2019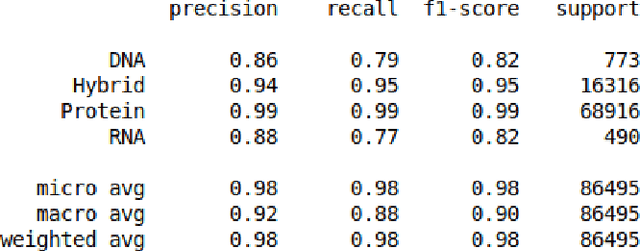
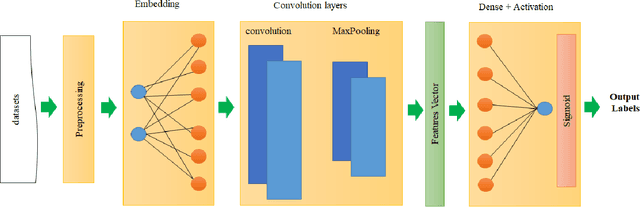
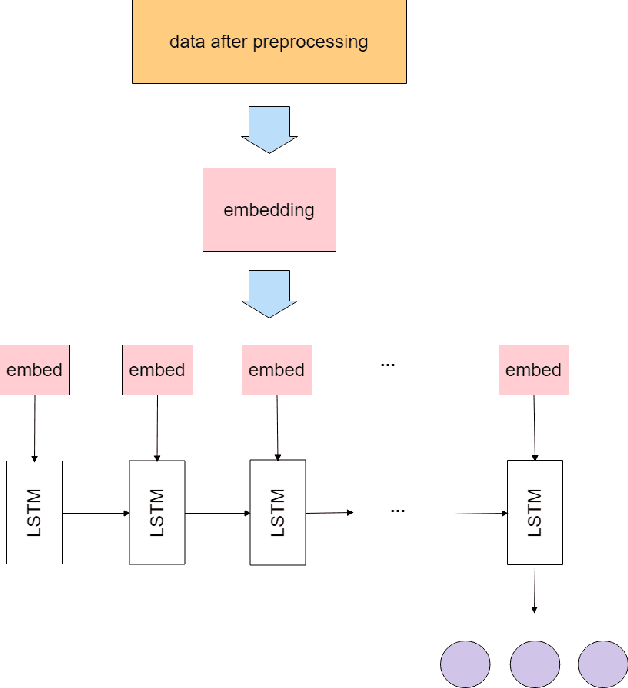
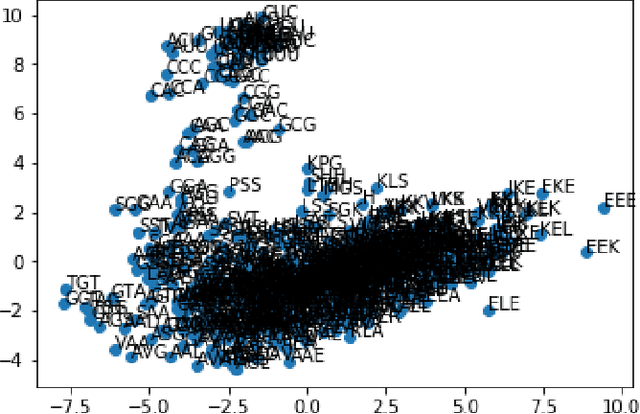
The study of the amino acid sequence is vital in life sciences. In this paper, we are using deep learning to solve macromolecule classification problem using amino acids. Deep learning has emerged as a strong and efficient framework that can be applied to a broad spectrum of complex learning problems which were difficult to solve using traditional machine learning techniques in the past. We are using word embedding from NLP to represent the amino acid sequence as vectors. We are using different deep learning model for classification of macromolecules like CNN, LSTM, and GRU. Convolution neural network can extract features from amino acid sequences which are represented by vectors. The extracted features will be feed to a different type of model to train a robust classifier. our results show that Word2vec as embedding combine with VGG-16 has better performance than LSTM and GRU. our approach gets an error rate of 1.5%. Code is available at https://github.com/say2sarwar/DeepAcid
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge