Sarfaraz Masood
Parkinson's Disease Diagnosis Through Deep Learning: A Novel LSTM-Based Approach for Freezing of Gait Detection
Dec 09, 2024Abstract:Deep learning holds tremendous potential in healthcare for uncovering hidden patterns within extensive clinical datasets, aiding in the diagnosis of various diseases. Parkinson's disease (PD) is a neurodegenerative condition characterized by the deterioration of brain function. In the initial stages of PD, automatic diagnosis poses a challenge due to the similarity in behavior between individuals with PD and those who are healthy. Our objective is to propose an effective model that can aid in the early detection of Parkinson's disease. We employed the VGRF gait signal dataset sourced from Physionet for distinguishing between healthy individuals and those diagnosed with Parkinson's disease. This paper introduces a novel deep learning architecture based on the LSTM network for automatically detecting freezing of gait episodes in Parkinson's disease patients. In contrast to conventional machine learning algorithms, this method eliminates manual feature engineering and proficiently captures prolonged temporal dependencies in gait patterns, thereby improving the diagnosis of Parkinson's disease. The LSTM network resolves the issue of vanishing gradients by employing memory blocks in place of self-connected hidden units, allowing for optimal information assimilation. To prevent overfitting, dropout and L2 regularization techniques have been employed. Additionally, the stochastic gradient-based optimizer Adam is used for the optimization process. The results indicate that our proposed approach surpasses current state-of-the-art models in FOG episode detection, achieving an accuracy of 97.71%, sensitivity of 99%, precision of 98%, and specificity of 96%. This demonstrates its potential as a superior classification method for Parkinson's disease detection.
Exploring learning environments for label\-efficient cancer diagnosis
Aug 15, 2024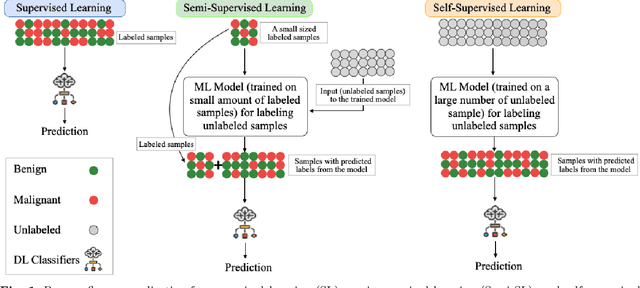
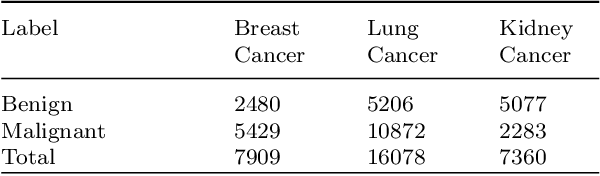
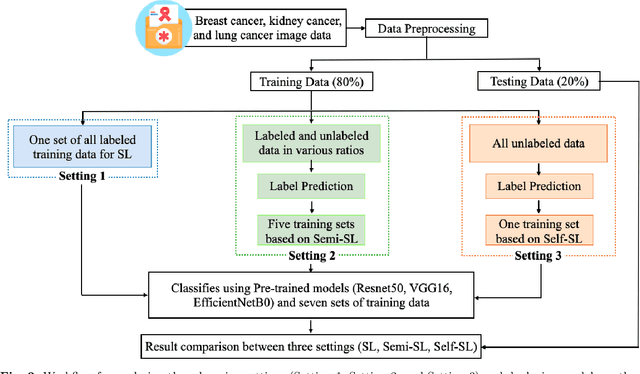
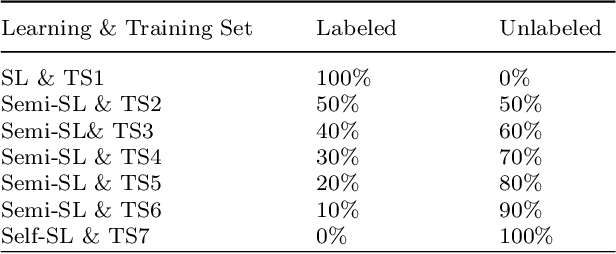
Abstract:Despite significant research efforts and advancements, cancer remains a leading cause of mortality. Early cancer prediction has become a crucial focus in cancer research to streamline patient care and improve treatment outcomes. Manual tumor detection by histopathologists can be time consuming, prompting the need for computerized methods to expedite treatment planning. Traditional approaches to tumor detection rely on supervised learning, necessitates a large amount of annotated data for model training. However, acquiring such extensive labeled data can be laborious and time\-intensive. This research examines the three learning environments: supervised learning (SL), semi\-supervised learning (Semi\-SL), and self\-supervised learning (Self\-SL): to predict kidney, lung, and breast cancer. Three pre\-trained deep learning models (Residual Network\-50, Visual Geometry Group\-16, and EfficientNetB0) are evaluated based on these learning settings using seven carefully curated training sets. To create the first training set (TS1), SL is applied to all annotated image samples. Five training sets (TS2\-TS6) with different ratios of labeled and unlabeled cancer images are used to evaluateSemi\-SL. Unlabeled cancer images from the final training set (TS7) are utilized for Self\-SL assessment. Among different learning environments, outcomes from the Semi\-SL setting show a strong degree of agreement with the outcomes achieved in the SL setting. The uniform pattern of observations from the pre\-trained models across all three datasets validates the methodology and techniques of the research. Based on modest number of labeled samples and minimal computing cost, our study suggests that the Semi\-SL option can be a highly viable replacement for the SL option under label annotation constraint scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge