Exploring learning environments for label\-efficient cancer diagnosis
Paper and Code
Aug 15, 2024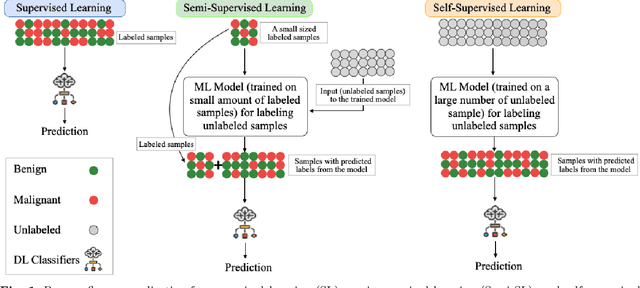
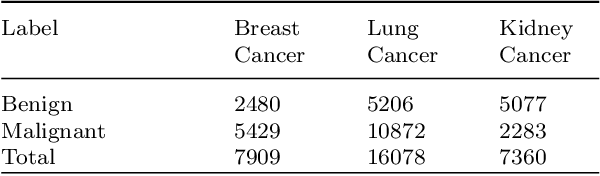
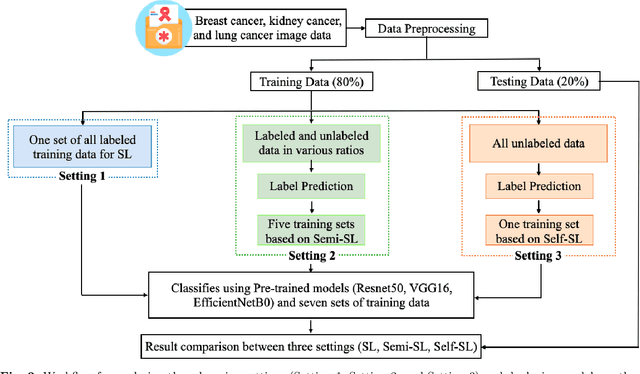
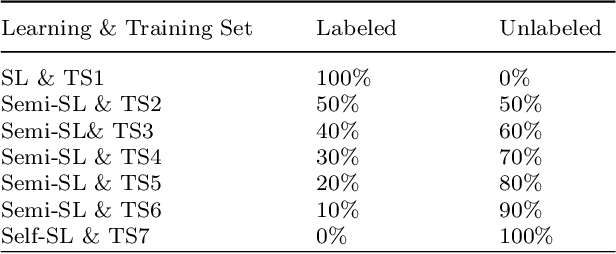
Despite significant research efforts and advancements, cancer remains a leading cause of mortality. Early cancer prediction has become a crucial focus in cancer research to streamline patient care and improve treatment outcomes. Manual tumor detection by histopathologists can be time consuming, prompting the need for computerized methods to expedite treatment planning. Traditional approaches to tumor detection rely on supervised learning, necessitates a large amount of annotated data for model training. However, acquiring such extensive labeled data can be laborious and time\-intensive. This research examines the three learning environments: supervised learning (SL), semi\-supervised learning (Semi\-SL), and self\-supervised learning (Self\-SL): to predict kidney, lung, and breast cancer. Three pre\-trained deep learning models (Residual Network\-50, Visual Geometry Group\-16, and EfficientNetB0) are evaluated based on these learning settings using seven carefully curated training sets. To create the first training set (TS1), SL is applied to all annotated image samples. Five training sets (TS2\-TS6) with different ratios of labeled and unlabeled cancer images are used to evaluateSemi\-SL. Unlabeled cancer images from the final training set (TS7) are utilized for Self\-SL assessment. Among different learning environments, outcomes from the Semi\-SL setting show a strong degree of agreement with the outcomes achieved in the SL setting. The uniform pattern of observations from the pre\-trained models across all three datasets validates the methodology and techniques of the research. Based on modest number of labeled samples and minimal computing cost, our study suggests that the Semi\-SL option can be a highly viable replacement for the SL option under label annotation constraint scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge