Sanghyeon Kim
Machine Learning Approach to Brain Tumor Detection and Classification
Oct 16, 2024



Abstract:Brain tumor detection and classification are critical tasks in medical image analysis, particularly in early-stage diagnosis, where accurate and timely detection can significantly improve treatment outcomes. In this study, we apply various statistical and machine learning models to detect and classify brain tumors using brain MRI images. We explore a variety of statistical models including linear, logistic, and Bayesian regressions, and the machine learning models including decision tree, random forest, single-layer perceptron, multi-layer perceptron, convolutional neural network (CNN), recurrent neural network, and long short-term memory. Our findings show that CNN outperforms other models, achieving the best performance. Additionally, we confirm that the CNN model can also work for multi-class classification, distinguishing between four categories of brain MRI images such as normal, glioma, meningioma, and pituitary tumor images. This study demonstrates that machine learning approaches are suitable for brain tumor detection and classification, facilitating real-world medical applications in assisting radiologists with early and accurate diagnosis.
Hydra: Multi-head Low-rank Adaptation for Parameter Efficient Fine-tuning
Sep 13, 2023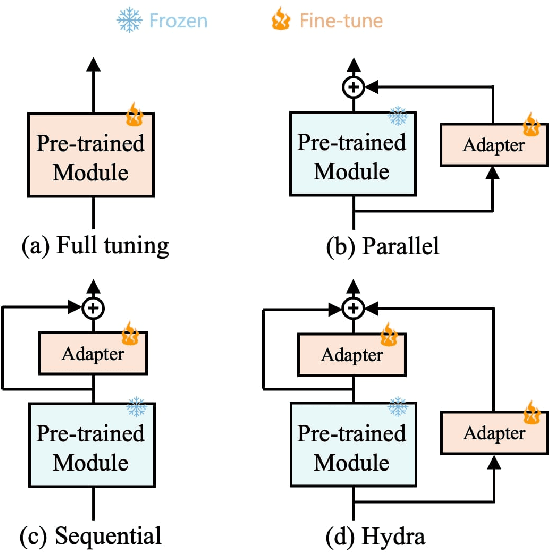
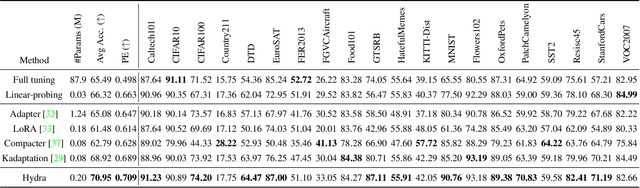

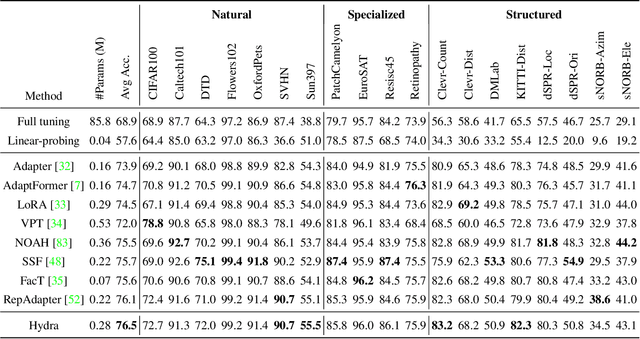
Abstract:The recent surge in large-scale foundation models has spurred the development of efficient methods for adapting these models to various downstream tasks. Low-rank adaptation methods, such as LoRA, have gained significant attention due to their outstanding parameter efficiency and no additional inference latency. This paper investigates a more general form of adapter module based on the analysis that parallel and sequential adaptation branches learn novel and general features during fine-tuning, respectively. The proposed method, named Hydra, due to its multi-head computational branches, combines parallel and sequential branch to integrate capabilities, which is more expressive than existing single branch methods and enables the exploration of a broader range of optimal points in the fine-tuning process. In addition, the proposed adaptation method explicitly leverages the pre-trained weights by performing a linear combination of the pre-trained features. It allows the learned features to have better generalization performance across diverse downstream tasks. Furthermore, we perform a comprehensive analysis of the characteristics of each adaptation branch with empirical evidence. Through an extensive range of experiments, encompassing comparisons and ablation studies, we substantiate the efficiency and demonstrate the superior performance of Hydra. This comprehensive evaluation underscores the potential impact and effectiveness of Hydra in a variety of applications. Our code is available on \url{https://github.com/extremebird/Hydra}
SMPConv: Self-moving Point Representations for Continuous Convolution
Apr 05, 2023Abstract:Continuous convolution has recently gained prominence due to its ability to handle irregularly sampled data and model long-term dependency. Also, the promising experimental results of using large convolutional kernels have catalyzed the development of continuous convolution since they can construct large kernels very efficiently. Leveraging neural networks, more specifically multilayer perceptrons (MLPs), is by far the most prevalent approach to implementing continuous convolution. However, there are a few drawbacks, such as high computational costs, complex hyperparameter tuning, and limited descriptive power of filters. This paper suggests an alternative approach to building a continuous convolution without neural networks, resulting in more computationally efficient and improved performance. We present self-moving point representations where weight parameters freely move, and interpolation schemes are used to implement continuous functions. When applied to construct convolutional kernels, the experimental results have shown improved performance with drop-in replacement in the existing frameworks. Due to its lightweight structure, we are first to demonstrate the effectiveness of continuous convolution in a large-scale setting, e.g., ImageNet, presenting the improvements over the prior arts. Our code is available on https://github.com/sangnekim/SMPConv
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge