Samuel J. Raymond
Towards Better Shale Gas Production Forecasting Using Transfer Learning
Jun 21, 2021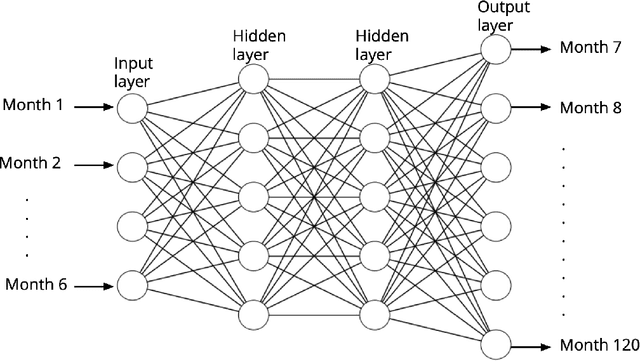
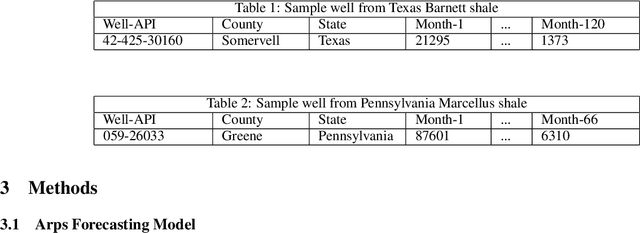

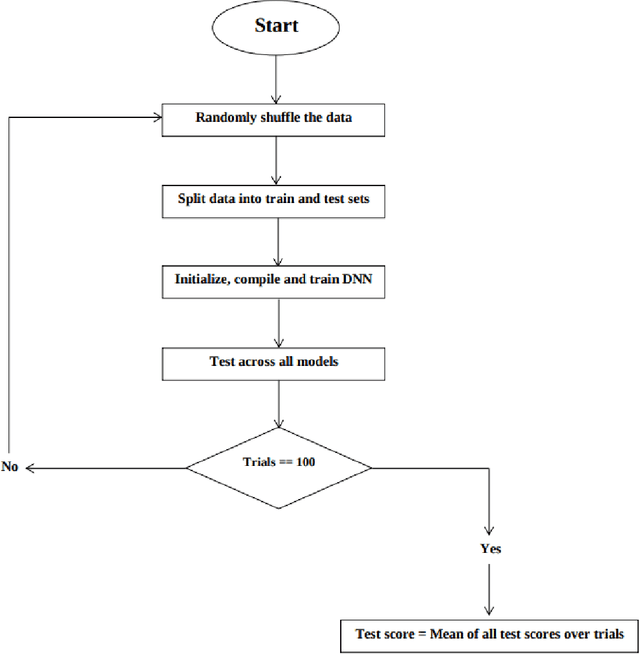
Abstract:Deep neural networks can generate more accurate shale gas production forecasts in counties with a limited number of sample wells by utilizing transfer learning. This paper provides a way of transferring the knowledge gained from other deep neural network models trained on adjacent counties into the county of interest. The paper uses data from more than 6000 shale gas wells across 17 counties from Texas Barnett and Pennsylvania Marcellus shale formations to test the capabilities of transfer learning. The results reduce the forecasting error between 11% and 47% compared to the widely used Arps decline curve model.
Applying physics-based loss functions to neural networks for improved generalizability in mechanics problems
May 25, 2021



Abstract:Physics-Informed Machine Learning (PIML) has gained momentum in the last 5 years with scientists and researchers aiming to utilize the benefits afforded by advances in machine learning, particularly in deep learning. With large scientific data sets with rich spatio-temporal data and high-performance computing providing large amounts of data to be inferred and interpreted, the task of PIML is to ensure that these predictions, categorizations, and inferences are enforced by, and conform to the limits imposed by physical laws. In this work a new approach to utilizing PIML is discussed that deals with the use of physics-based loss functions. While typical usage of physical equations in the loss function requires complex layers of derivatives and other functions to ensure that the known governing equation is satisfied, here we show that a similar level of enforcement can be found by implementing more simpler loss functions on specific kinds of output data. The generalizability that this approach affords is shown using examples of simple mechanical models that can be thought of as sufficiently simplified surrogate models for a wide class of problems.
Classification of head impacts based on the spectral density of measurable kinematics
Apr 19, 2021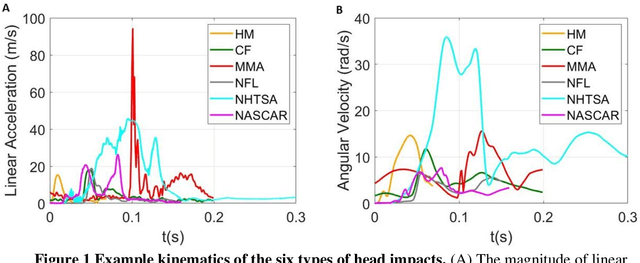
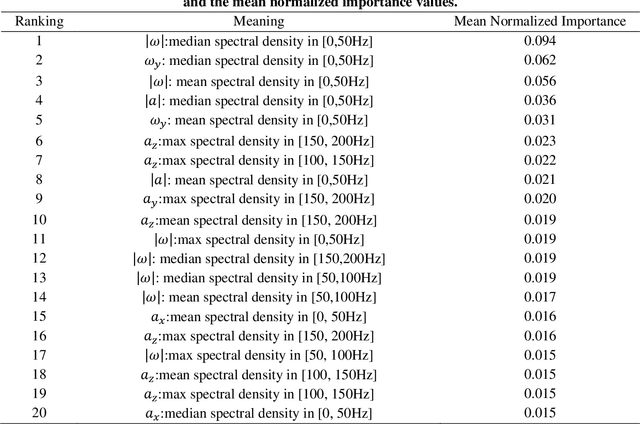
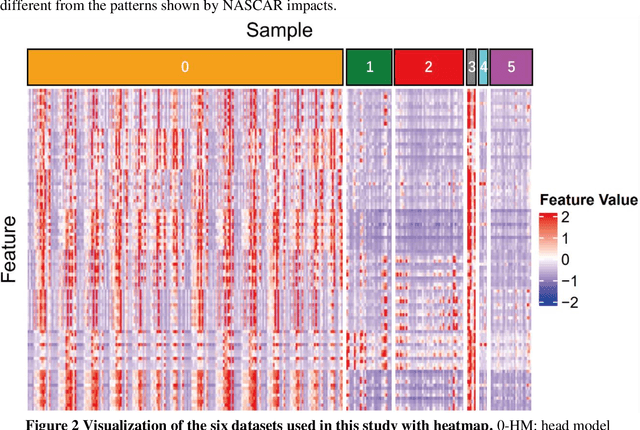
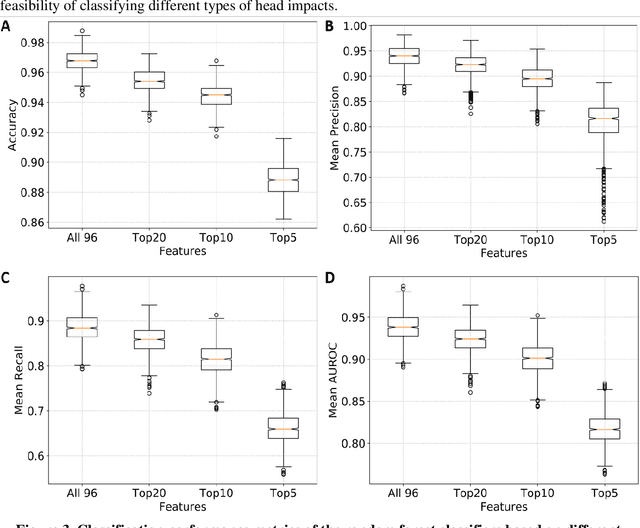
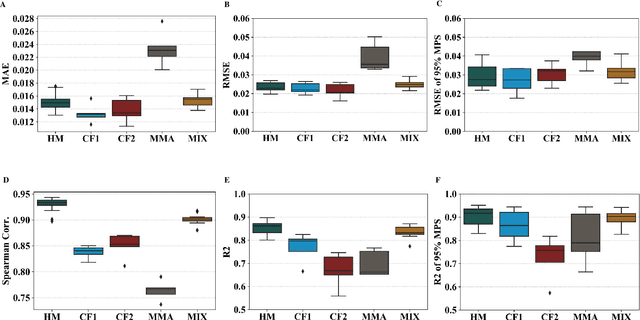
Abstract:Traumatic brain injury can be caused by head impacts, but many brain injury risk estimation models are less accurate across the variety of impacts that patients may undergo. In this study, we investigated the spectral characteristics of different head impact types with kinematics classification. Data was analyzed from 3262 head impacts from head model simulations, on-field data from American football and mixed martial arts (MMA) using our instrumented mouthguard, and publicly available car crash data. A random forest classifier with spectral densities of linear acceleration and angular velocity was built to classify different types of head impacts (e.g., football, MMA), reaching a median accuracy of 96% over 1000 random partitions of training and test sets. Furthermore, to test the classifier on data from different measurement devices, another 271 lab-reconstructed impacts were obtained from 5 other instrumented mouthguards with the classifier reaching over 96% accuracy from these devices. The most important features in classification included both low-frequency and high-frequency features, both linear acceleration features and angular velocity features. It was found that different head impact types had different distributions of spectral densities in low-frequency and high-frequency ranges (e.g., the spectral densities of MMA impacts were higher in high-frequency range than in the low-frequency range). Finally, with head impact classification, type-specific, nearest-neighbor regression models were built for 95th percentile maximum principal strain, 95th percentile maximum principal strain in corpus callosum, and cumulative strain damage (15th percentile). This showed a generally higher R^2-value than baseline models without classification.
Prediction of brain strain across head impact subtypes using 18 brain injury criteria
Dec 18, 2020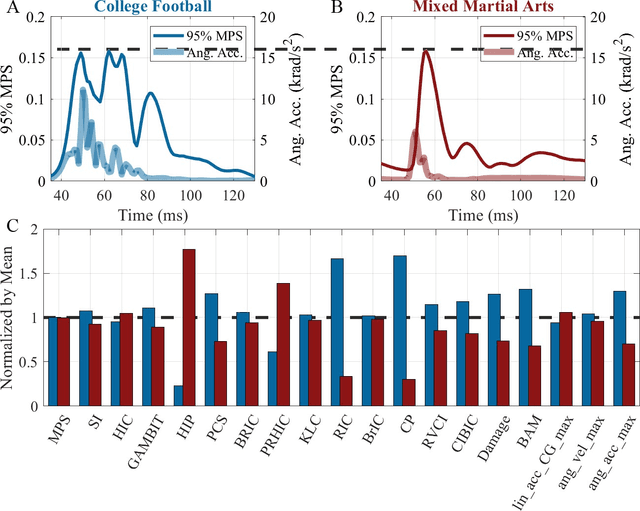
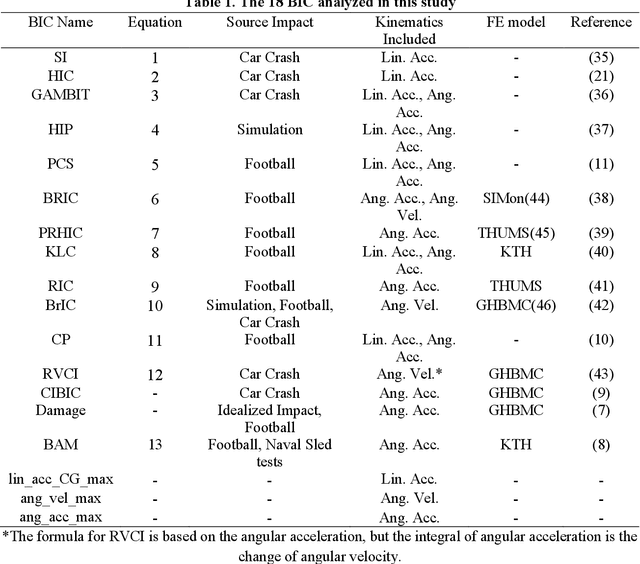
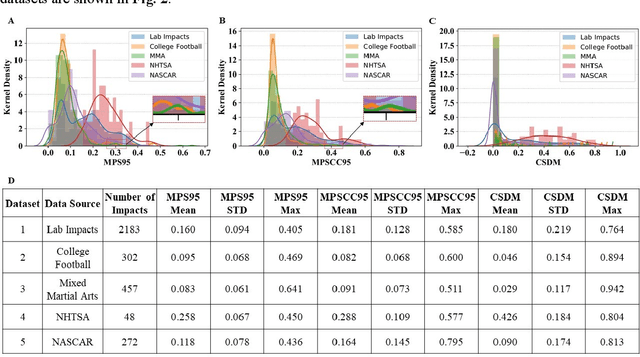

Abstract:Multiple brain injury criteria (BIC) are developed to quickly quantify brain injury risks after head impacts. These BIC originated from different types of head impacts (e.g., sports and car crashes) are widely used in risk evaluation. However, the predictability of the BIC on different types of head impacts has not been evaluated. Physiologically, the brain strain is often considered the key parameter of brain injury. To evaluate the BIC's ability to predict brain strain across five datasets comprising different head impact subtypes, linear regression was used to model 95% maximum principal strain, 95% maximum principal strain at corpus callosum, and cumulative strain damage (15%) on 18 BIC. The results show significant differences in the relationship between BIC and brain strain across datasets, indicating the same BIC value may indicate different brain strain in different head impact subtypes. The accuracy of regression is generally decreasing if the BIC regression models are fit on a dataset with a different head impact subtype rather than on the dataset with the same subtype. Given this finding, this study raises concerns for applying BIC to predict the brain strain for head impacts different from the head impacts on which the BIC was developed.
Deep Learning Head Model for Real-time Estimation of Entire Brain Deformation in Concussion
Oct 20, 2020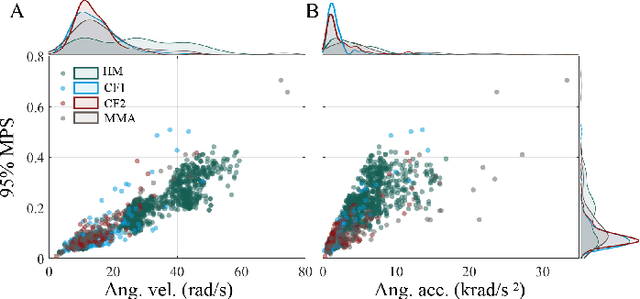
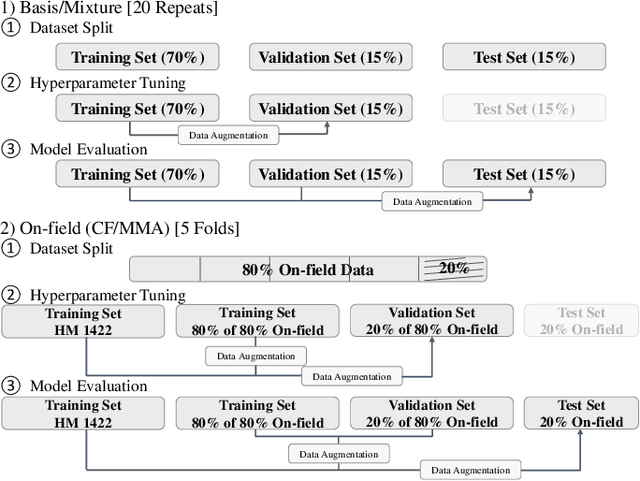

Abstract:Objective: Many recent studies have suggested that brain deformation resulting from a head impact is linked to the corresponding clinical outcome, such as mild traumatic brain injury (mTBI). Even though several finite element (FE) head models have been developed and validated to calculate brain deformation based on impact kinematics, the clinical application of these FE head models is limited due to the time-consuming nature of FE simulations. This work aims to accelerate the process of brain deformation calculation and thus improve the potential for clinical applications. Methods: We propose a deep learning head model with a five-layer deep neural network and feature engineering, and trained and tested the model on 1803 total head impacts from a combination of head model simulations and on-field college football and mixed martial arts impacts. Results: The proposed deep learning head model can calculate the maximum principal strain for every element in the entire brain in less than 0.001s (with an average root mean squared error of 0.025, and with a standard deviation of 0.002 over twenty repeats with random data partition and model initialization). The contributions of various features to the predictive power of the model were investigated, and it was noted that the features based on angular acceleration were found to be more predictive than the features based on angular velocity. Conclusion: Trained using the dataset of 1803 head impacts, this model can be applied to various sports in the calculation of brain strain with accuracy, and its applicability can even further be extended by incorporating data from other types of head impacts. Significance: In addition to the potential clinical application in real-time brain deformation monitoring, this model will help researchers estimate the brain strain from a large number of head impacts more efficiently than using FE models.
A New Open-Access Platform for Measuring and Sharing mTBI Data
Oct 16, 2020
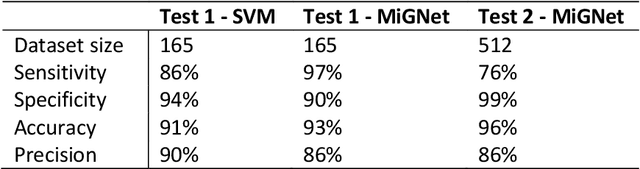
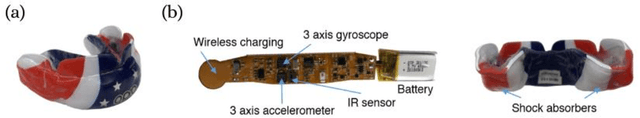
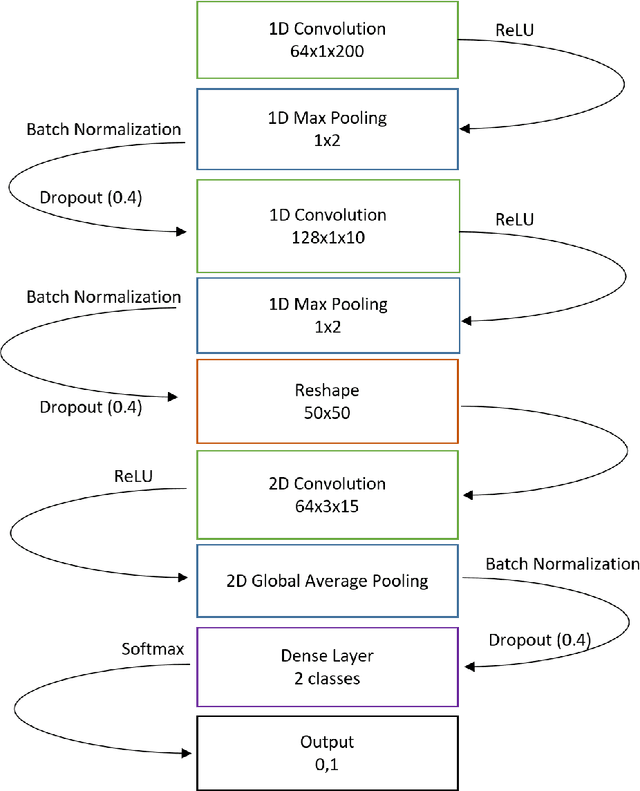
Abstract:Despite numerous research efforts, the precise mechanisms of concussion have yet to be fully uncovered. Clinical studies on high-risk populations, such as contact sports athletes, have become more common and give insight on the link between impact severity and brain injury risk through the use of wearable sensors and neurological testing. However, as the number of institutions operating these studies grows, there is a growing need for a platform to share these data to facilitate our understanding of concussion mechanisms and aid in the development of suitable diagnostic tools. To that end, this paper puts forth two contributions: 1) a centralized, open-source platform for storing and sharing head impact data, in collaboration with the Federal Interagency Traumatic Brain Injury Research informatics system (FITBIR), and 2) a deep learning impact detection algorithm (MiGNet) to differentiate between true head impacts and false positives for the previously biomechanically validated instrumented mouthguard sensor (MiG2.0), all of which easily interfaces with FITBIR. We report 96% accuracy using MiGNet, based on a neural network model, improving on previous work based on Support Vector Machines achieving 91% accuracy, on an out of sample dataset of high school and collegiate football head impacts. The integrated MiG2.0 and FITBIR system serve as a collaborative research tool to be disseminated across multiple institutions towards creating a standardized dataset for furthering the knowledge of concussion biomechanics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge