Rustam Zhumagambetov
Explainable AI needs formal notions of explanation correctness
Sep 26, 2024Abstract:The use of machine learning (ML) in critical domains such as medicine poses risks and requires regulation. One requirement is that decisions of ML systems in high-risk applications should be human-understandable. The field of "explainable artificial intelligence" (XAI) seemingly addresses this need. However, in its current form, XAI is unfit to provide quality control for ML; it itself needs scrutiny. Popular XAI methods cannot reliably answer important questions about ML models, their training data, or a given test input. We recapitulate results demonstrating that popular XAI methods systematically attribute importance to input features that are independent of the prediction target. This limits their utility for purposes such as model and data (in)validation, model improvement, and scientific discovery. We argue that the fundamental reason for this limitation is that current XAI methods do not address well-defined problems and are not evaluated against objective criteria of explanation correctness. Researchers should formally define the problems they intend to solve first and then design methods accordingly. This will lead to notions of explanation correctness that can be theoretically verified and objective metrics of explanation performance that can be assessed using ground-truth data.
A biologically-inspired evaluation of molecular generative machine learning
Aug 20, 2022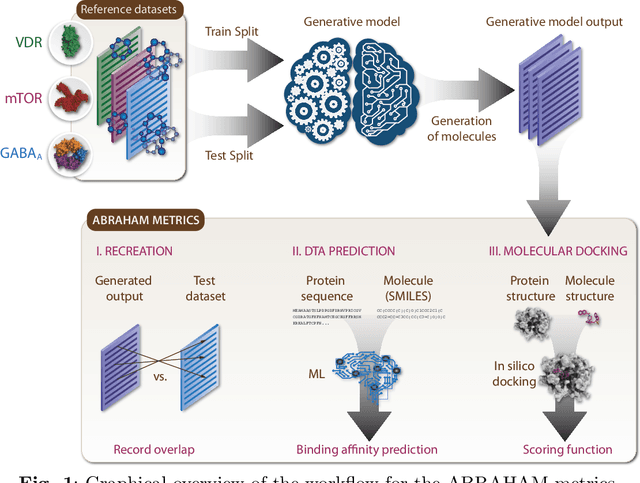
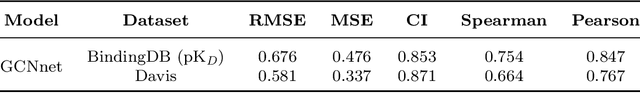
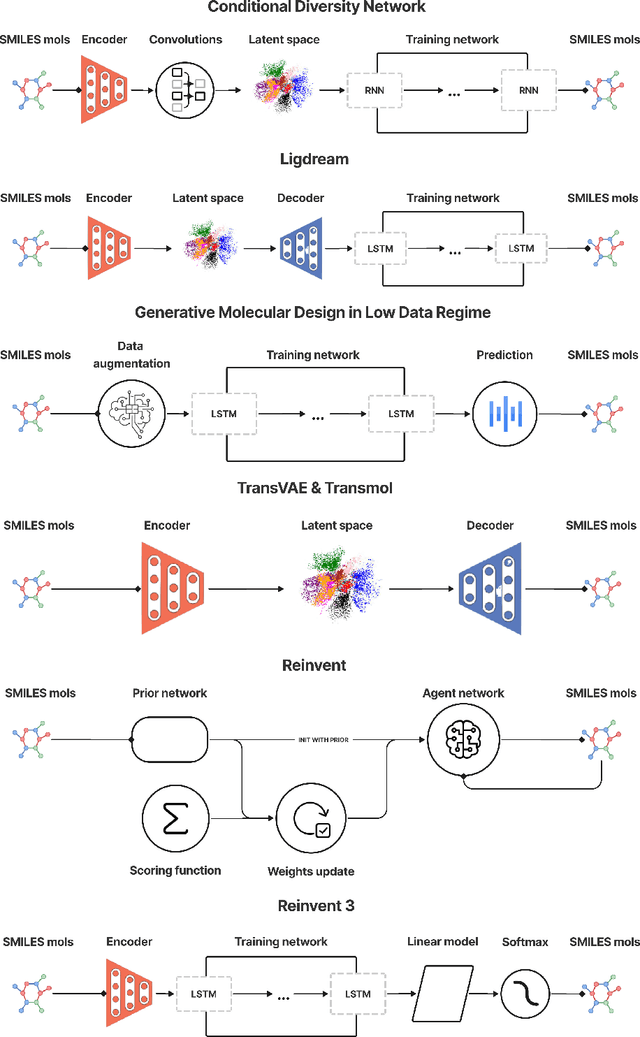
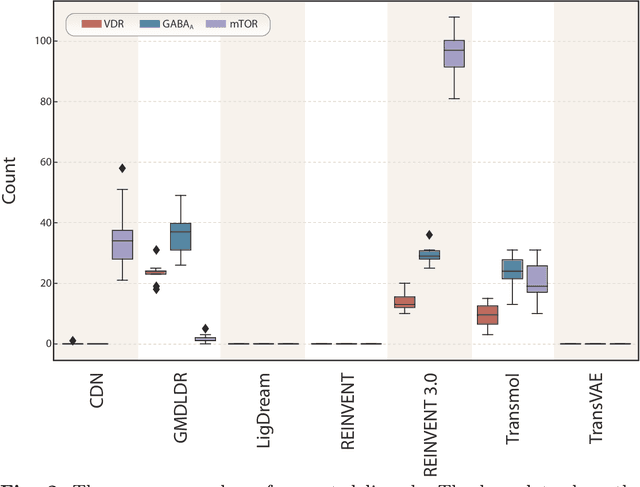
Abstract:While generative models have recently become ubiquitous in many scientific areas, less attention has been paid to their evaluation. For molecular generative models, the state-of-the-art examines their output in isolation or in relation to its input. However, their biological and functional properties, such as ligand-target interaction is not being addressed. In this study, a novel biologically-inspired benchmark for the evaluation of molecular generative models is proposed. Specifically, three diverse reference datasets are designed and a set of metrics are introduced which are directly relevant to the drug discovery process. In particular we propose a recreation metric, apply drug-target affinity prediction and molecular docking as complementary techniques for the evaluation of generative outputs. While all three metrics show consistent results across the tested generative models, a more detailed comparison of drug-target affinity binding and molecular docking scores revealed that unimodal predictiors can lead to erroneous conclusions about target binding on a molecular level and a multi-modal approach is thus preferrable. The key advantage of this framework is that it incorporates prior physico-chemical domain knowledge into the benchmarking process by focusing explicitly on ligand-target interactions and thus creating a highly efficient tool not only for evaluating molecular generative outputs in particular, but also for enriching the drug discovery process in general.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge