Russ Greiner
FraGNNet: A Deep Probabilistic Model for Mass Spectrum Prediction
Apr 02, 2024



Abstract:The process of identifying a compound from its mass spectrum is a critical step in the analysis of complex mixtures. Typical solutions for the mass spectrum to compound (MS2C) problem involve matching the unknown spectrum against a library of known spectrum-molecule pairs, an approach that is limited by incomplete library coverage. Compound to mass spectrum (C2MS) models can improve retrieval rates by augmenting real libraries with predicted spectra. Unfortunately, many existing C2MS models suffer from problems with prediction resolution, scalability, or interpretability. We develop a new probabilistic method for C2MS prediction, FraGNNet, that can efficiently and accurately predict high-resolution spectra. FraGNNet uses a structured latent space to provide insight into the underlying processes that define the spectrum. Our model achieves state-of-the-art performance in terms of prediction error, and surpasses existing C2MS models as a tool for retrieval-based MS2C.
Ischemic Stroke Lesion Prediction using imbalanced Temporal Deep Gaussian Process (iTDGP)
Nov 16, 2022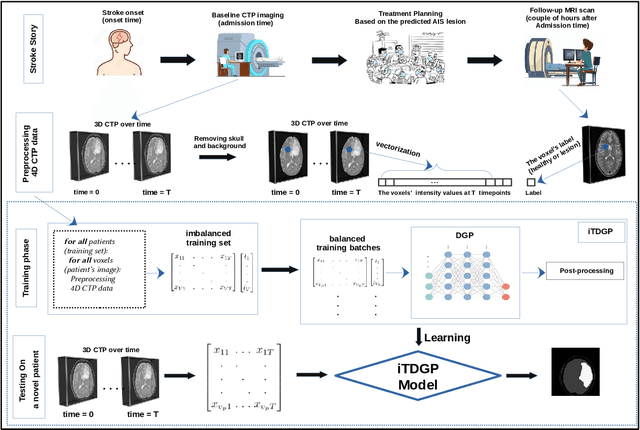
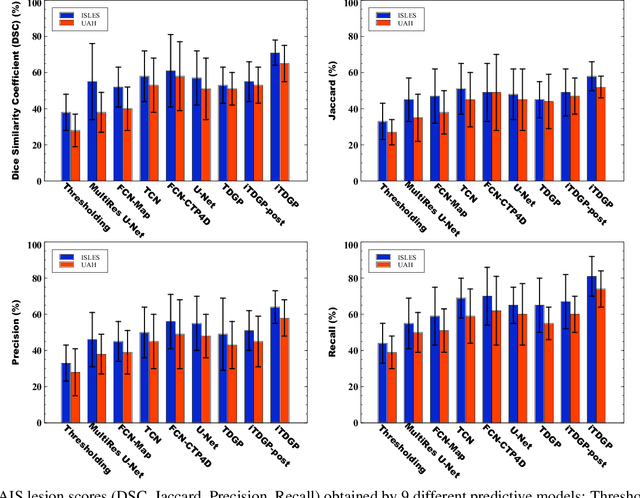
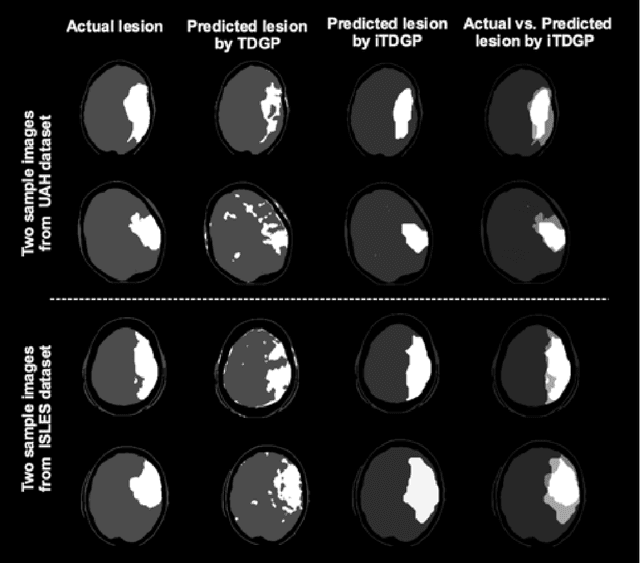
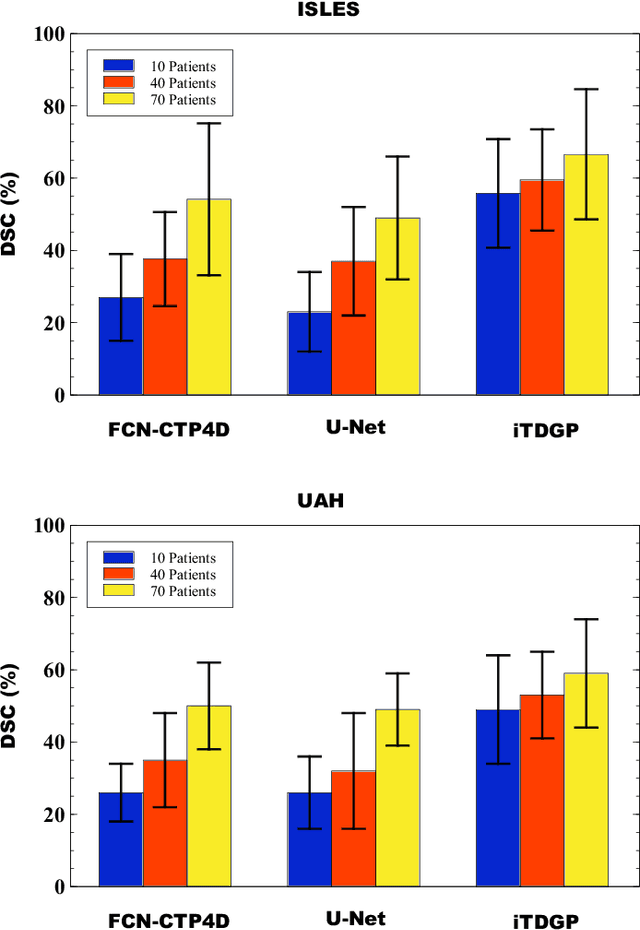
Abstract:As one of the leading causes of mortality and disability worldwide, Acute Ischemic Stroke (AIS) occurs when the blood supply to the brain is suddenly interrupted because of a blocked artery. Within seconds of AIS onset, the brain cells surrounding the blocked artery die, which leads to the progression of the lesion. The automated and precise prediction of the existing lesion plays a vital role in the AIS treatment planning and prevention of further injuries. The current standard AIS assessment method, which thresholds the 3D measurement maps extracted from Computed Tomography Perfusion (CTP) images, is not accurate enough. Due to this fact, in this article, we propose the imbalanced Temporal Deep Gaussian Process (iTDGP), a probabilistic model that can improve AIS lesions prediction by using baseline CTP time series. Our proposed model can effectively extract temporal information from the CTP time series and map it to the class labels of the brain's voxels. In addition, by using batch training and voxel-level analysis iTDGP can learn from a few patients and it is robust against imbalanced classes. Moreover, our model incorporates a post-processor capable of improving prediction accuracy using spatial information. Our comprehensive experiments, on the ISLES 2018 and the University of Alberta Hospital (UAH) datasets, show that iTDGP performs better than state-of-the-art AIS lesion predictors, obtaining the (cross-validation) Dice score of 71.42% and 65.37% with a significant p<0.05, respectively.
Predicting the Long-Term Outcomes of Biologics in Psoriasis Patients Using Machine Learning
Aug 25, 2019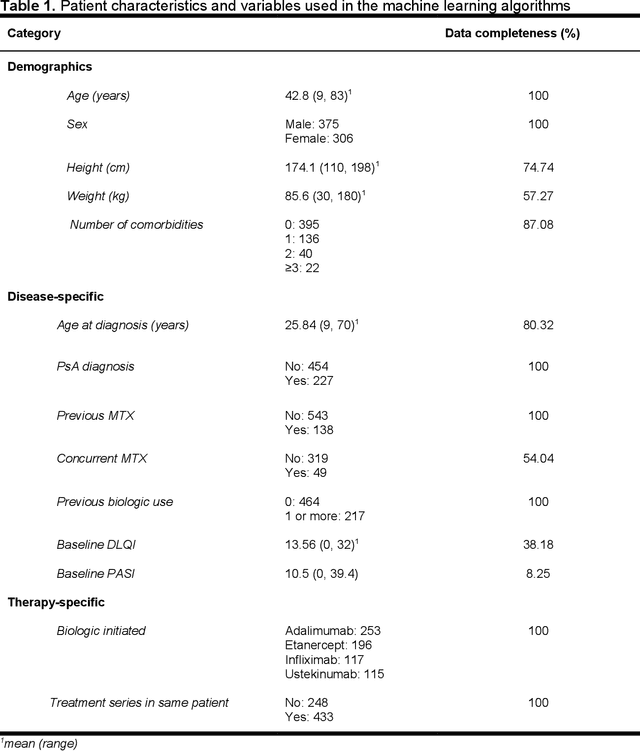
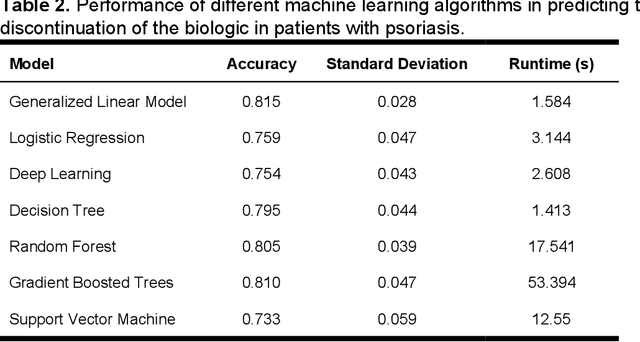
Abstract:Background. Real-world data show that approximately 50% of psoriasis patients treated with a biologic agent will discontinue the drug because of loss of efficacy. History of previous therapy with another biologic, female sex and obesity were identified as predictors of drug discontinuations, but their individual predictive value is low. Objectives. To determine whether machine learning algorithms can produce models that can accurately predict outcomes of biologic therapy in psoriasis on individual patient level. Results. All tested machine learning algorithms could accurately predict the risk of drug discontinuation and its cause (e.g. lack of efficacy vs adverse event). The learned generalized linear model achieved diagnostic accuracy of 82%, requiring under 2 seconds per patient using the psoriasis patients dataset. Input optimization analysis established a profile of a patient who has best chances of long-term treatment success: biologic-naive patient under 49 years, early-onset plaque psoriasis without psoriatic arthritis, weight < 100 kg, and moderate-to-severe psoriasis activity (DLQI $\geq$ 16; PASI $\geq$ 10). Moreover, a different generalized linear model is used to predict the length of treatment for each patient with mean absolute error (MAE) of 4.5 months. However Pearson Correlation Coefficient indicates 0.935 linear dependencies between the actual treatment lengths and predicted ones. Conclusions. Machine learning algorithms predict the risk of drug discontinuation and treatment duration with accuracy exceeding 80%, based on a small set of predictive variables. This approach can be used as a decision-making tool, communicating expected outcomes to the patient, and development of evidence-based guidelines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge