Ruchira Basu
Target specific peptide design using latent space approximate trajectory collector
Feb 02, 2023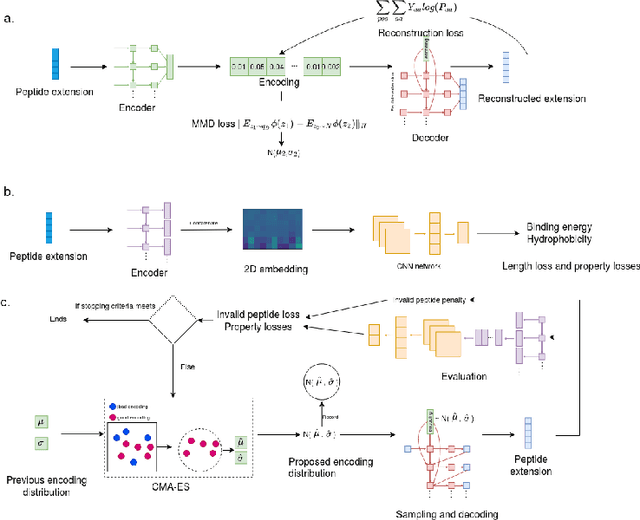

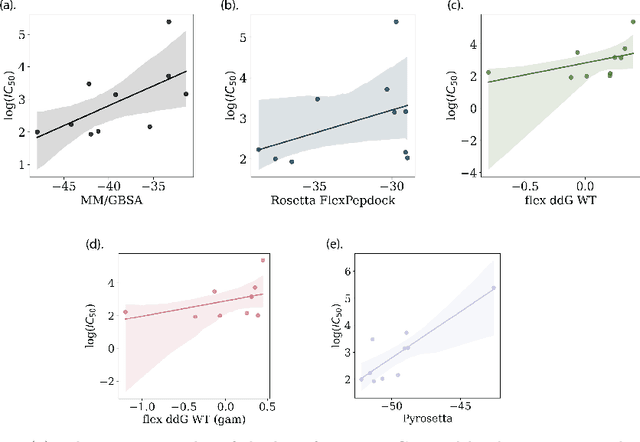
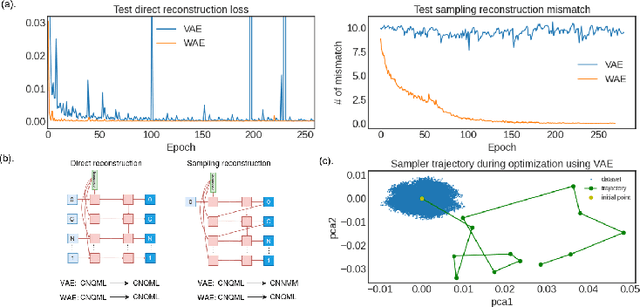
Abstract:Despite the prevalence and many successes of deep learning applications in de novo molecular design, the problem of peptide generation targeting specific proteins remains unsolved. A main barrier for this is the scarcity of the high-quality training data. To tackle the issue, we propose a novel machine learning based peptide design architecture, called Latent Space Approximate Trajectory Collector (LSATC). It consists of a series of samplers on an optimization trajectory on a highly non-convex energy landscape that approximates the distributions of peptides with desired properties in a latent space. The process involves little human intervention and can be implemented in an end-to-end manner. We demonstrate the model by the design of peptide extensions targeting Beta-catenin, a key nuclear effector protein involved in canonical Wnt signalling. When compared with a random sampler, LSATC can sample peptides with $36\%$ lower binding scores in a $16$ times smaller interquartile range (IQR) and $284\%$ less hydrophobicity with a $1.4$ times smaller IQR. LSATC also largely outperforms other common generative models. Finally, we utilized a clustering algorithm to select 4 peptides from the 100 LSATC designed peptides for experimental validation. The result confirms that all the four peptides extended by LSATC show improved Beta-catenin binding by at least $20.0\%$, and two of the peptides show a $3$ fold increase in binding affinity as compared to the base peptide.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge