Robert Azencott
Fast $k$-means clustering in Riemannian manifolds via Fréchet maps: Applications to large-dimensional SPD matrices
Nov 12, 2025Abstract:We introduce a novel, efficient framework for clustering data on high-dimensional, non-Euclidean manifolds that overcomes the computational challenges associated with standard intrinsic methods. The key innovation is the use of the $p$-Fréchet map $F^p : \mathcal{M} \to \mathbb{R}^\ell$ -- defined on a generic metric space $\mathcal{M}$ -- which embeds the manifold data into a lower-dimensional Euclidean space $\mathbb{R}^\ell$ using a set of reference points $\{r_i\}_{i=1}^\ell$, $r_i \in \mathcal{M}$. Once embedded, we can efficiently and accurately apply standard Euclidean clustering techniques such as k-means. We rigorously analyze the mathematical properties of $F^p$ in the Euclidean space and the challenging manifold of $n \times n$ symmetric positive definite matrices $\mathit{SPD}(n)$. Extensive numerical experiments using synthetic and real $\mathit{SPD}(n)$ data demonstrate significant performance gains: our method reduces runtime by up to two orders of magnitude compared to intrinsic manifold-based approaches, all while maintaining high clustering accuracy, including scenarios where existing alternative methods struggle or fail.
Automatic classification of deformable shapes
Nov 04, 2022Abstract:Let $\mathcal{D}$ be a dataset of smooth 3D-surfaces, partitioned into disjoint classes $\mathit{CL}_j$, $j= 1, \ldots, k$. We show how optimized diffeomorphic registration applied to large numbers of pairs $S,S' \in \mathcal{D}$ can provide descriptive feature vectors to implement automatic classification on $\mathcal{D}$, and generate classifiers invariant by rigid motions in $\mathbb{R}^3$. To enhance accuracy of automatic classification, we enrich the smallest classes $\mathit{CL}_j$ by diffeomorphic interpolation of smooth surfaces between pairs $S,S' \in \mathit{CL}_j$. We also implement small random perturbations of surfaces $S\in \mathit{CL}_j$ by random flows of smooth diffeomorphisms $F_t:\mathbb{R}^3 \to \mathbb{R}^3$. Finally, we test our automatic classification methods on a cardiology data base of discretized mitral valve surfaces.
Stochastic Neural Networks for Automatic Cell Tracking in Microscopy Image Sequences of Bacterial Colonies
Apr 27, 2021Abstract:We describe an automated analysis method to quantify the detailed growth dynamics of a population of bacilliform bacteria. We propose an innovative approach to frame-sequence tracking of deformable-cell motion by the automated minimization of a new, specific cost functional. This minimization is implemented by dedicated Boltzmann machines (stochastic recurrent neural networks). Automated detection of cell divisions is handled similarly by successive minimizations of two cost functions, alternating the identification of children pairs and parent identification. We validate this automatic cell tracking algorithm using recordings of simulated cell colonies that closely mimic the growth dynamics of \emph{E. coli} in microfluidic traps. On a batch of 1100 image frames, cell registration accuracies per frame ranged from 94.5\% to 100\%, with a high average. Our initial tests using experimental image sequences of \emph{E. coli} colonies also yield convincing results, with a registration accuracy ranging from 90\% to 100\%.
Markov Random Fields and Mass Spectra Discrimination
Oct 13, 2014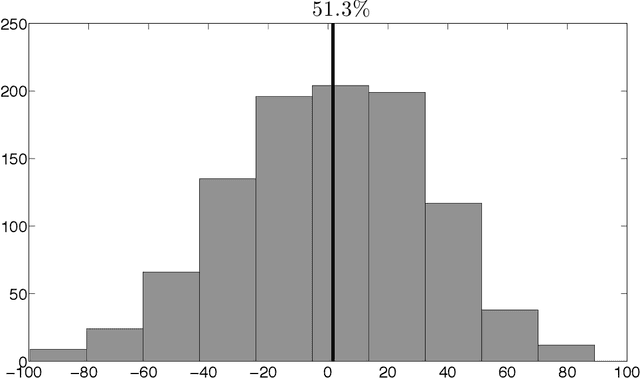
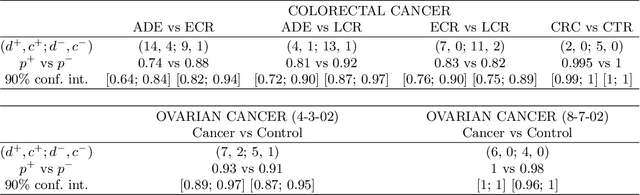
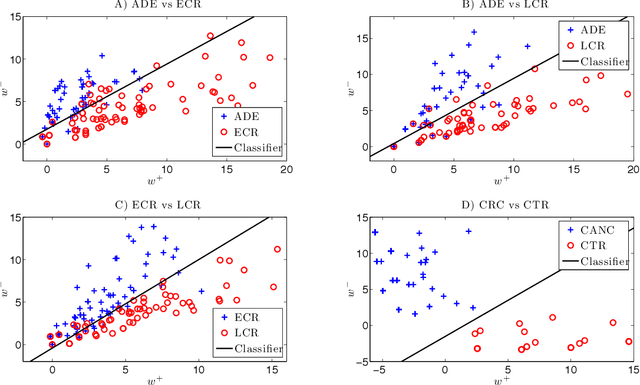
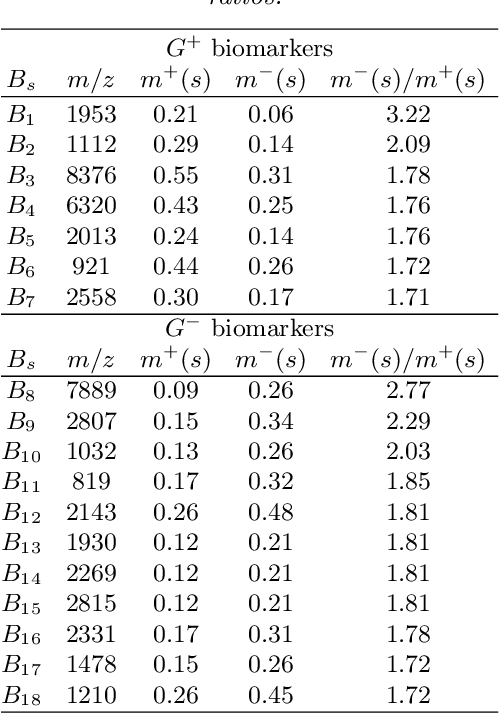
Abstract:For mass spectra acquired from cancer patients by MALDI or SELDI techniques, automated discrimination between cancer types or stages has often been implemented by machine learnings. These techniques typically generate "black-box" classifiers, which are difficult to interpret biologically. We develop new and efficient signature discovery algorithms leading to interpretable signatures combining the discriminating power of explicitly selected small groups of biomarkers, identified by their m/z ratios. Our approach is based on rigorous stochastic modeling of "homogeneous" datasets of mass spectra by a versatile class of parameterized Markov Random Fields. We present detailed algorithms validated by precise theoretical results. We also outline the successful tests of our approach to generate efficient explicit signatures for six benchmark discrimination tasks, based on mass spectra acquired from colorectal cancer patients, as well as from ovarian cancer patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge