Markov Random Fields and Mass Spectra Discrimination
Paper and Code
Oct 13, 2014
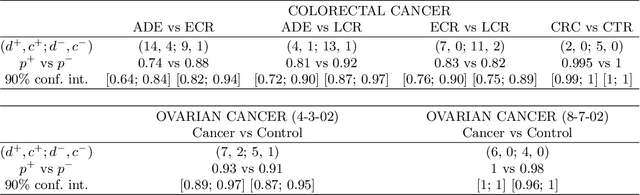
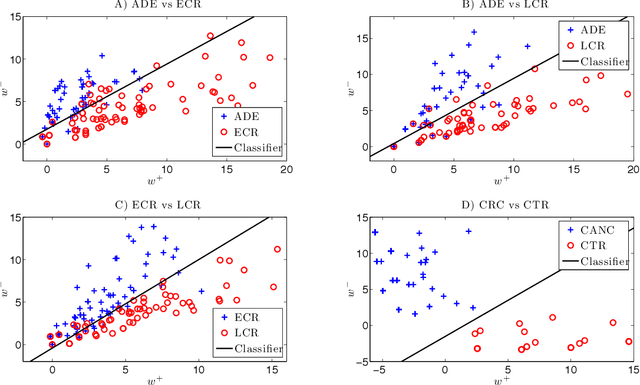
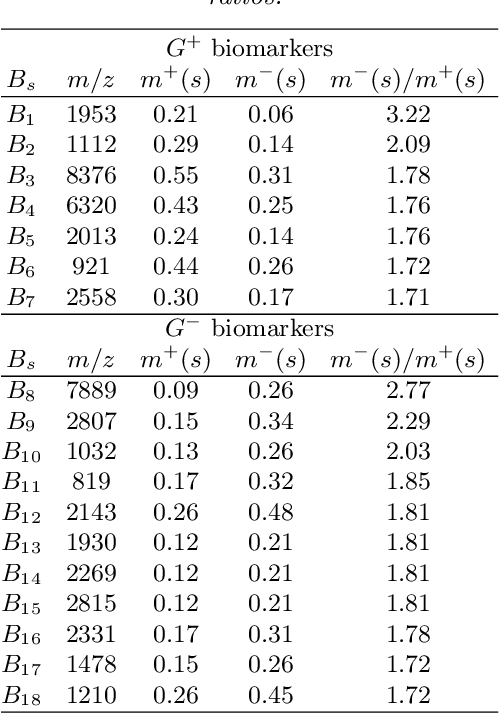
For mass spectra acquired from cancer patients by MALDI or SELDI techniques, automated discrimination between cancer types or stages has often been implemented by machine learnings. These techniques typically generate "black-box" classifiers, which are difficult to interpret biologically. We develop new and efficient signature discovery algorithms leading to interpretable signatures combining the discriminating power of explicitly selected small groups of biomarkers, identified by their m/z ratios. Our approach is based on rigorous stochastic modeling of "homogeneous" datasets of mass spectra by a versatile class of parameterized Markov Random Fields. We present detailed algorithms validated by precise theoretical results. We also outline the successful tests of our approach to generate efficient explicit signatures for six benchmark discrimination tasks, based on mass spectra acquired from colorectal cancer patients, as well as from ovarian cancer patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge