Ricky Molén
Efficient Mixture Learning in Black-Box Variational Inference
Jun 11, 2024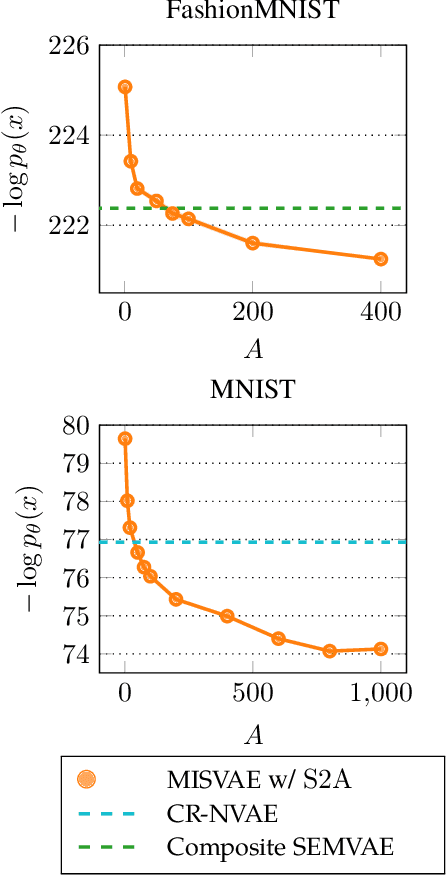
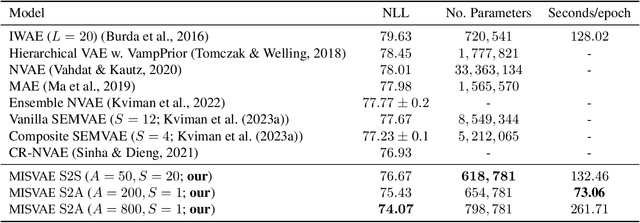
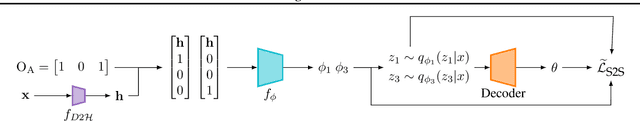
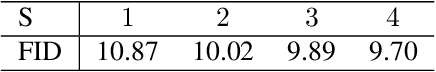
Abstract:Mixture variational distributions in black box variational inference (BBVI) have demonstrated impressive results in challenging density estimation tasks. However, currently scaling the number of mixture components can lead to a linear increase in the number of learnable parameters and a quadratic increase in inference time due to the evaluation of the evidence lower bound (ELBO). Our two key contributions address these limitations. First, we introduce the novel Multiple Importance Sampling Variational Autoencoder (MISVAE), which amortizes the mapping from input to mixture-parameter space using one-hot encodings. Fortunately, with MISVAE, each additional mixture component incurs a negligible increase in network parameters. Second, we construct two new estimators of the ELBO for mixtures in BBVI, enabling a tremendous reduction in inference time with marginal or even improved impact on performance. Collectively, our contributions enable scalability to hundreds of mixture components and provide superior estimation performance in shorter time, with fewer network parameters compared to previous Mixture VAEs. Experimenting with MISVAE, we achieve astonishing, SOTA results on MNIST. Furthermore, we empirically validate our estimators in other BBVI settings, including Bayesian phylogenetic inference, where we improve inference times for the SOTA mixture model on eight data sets.
Improved Variational Bayesian Phylogenetic Inference using Mixtures
Oct 02, 2023Abstract:We present VBPI-Mixtures, an algorithm designed to enhance the accuracy of phylogenetic posterior distributions, particularly for tree-topology and branch-length approximations. Despite the Variational Bayesian Phylogenetic Inference (VBPI), a leading-edge black-box variational inference (BBVI) framework, achieving remarkable approximations of these distributions, the multimodality of the tree-topology posterior presents a formidable challenge to sampling-based learning techniques such as BBVI. Advanced deep learning methodologies such as normalizing flows and graph neural networks have been explored to refine the branch-length posterior approximation, yet efforts to ameliorate the posterior approximation over tree topologies have been lacking. Our novel VBPI-Mixtures algorithm bridges this gap by harnessing the latest breakthroughs in mixture learning within the BBVI domain. As a result, VBPI-Mixtures is capable of capturing distributions over tree-topologies that VBPI fails to model. We deliver state-of-the-art performance on difficult density estimation tasks across numerous real phylogenetic datasets.
Learning with MISELBO: The Mixture Cookbook
Sep 30, 2022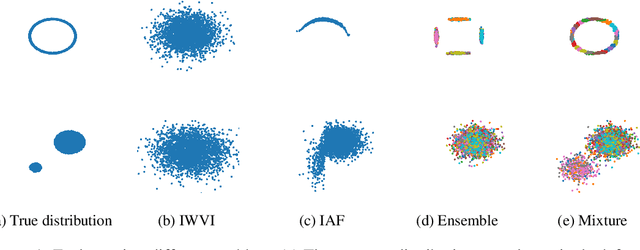
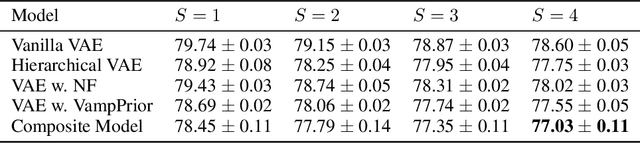
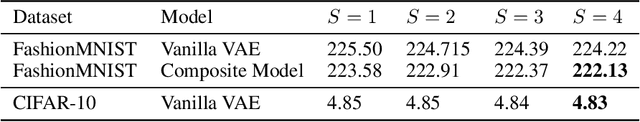
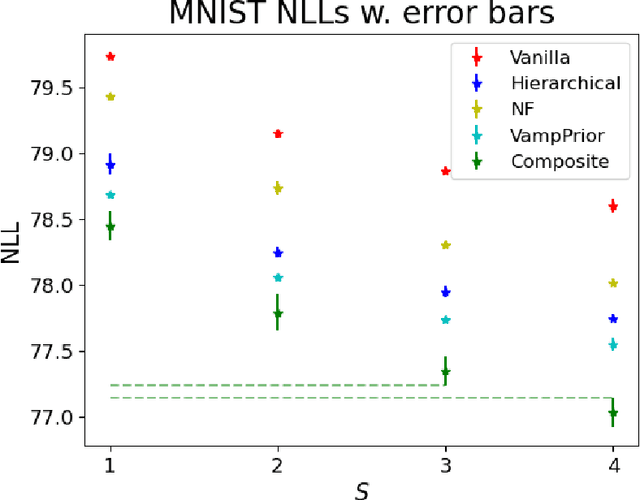
Abstract:Mixture models in variational inference (VI) is an active field of research. Recent works have established their connection to multiple importance sampling (MIS) through the MISELBO and advanced the use of ensemble approximations for large-scale problems. However, as we show here, an independent learning of the ensemble components can lead to suboptimal diversity. Hence, we study the effect of instead using MISELBO as an objective function for learning mixtures, and we propose the first ever mixture of variational approximations for a normalizing flow-based hierarchical variational autoencoder (VAE) with VampPrior and a PixelCNN decoder network. Two major insights led to the construction of this novel composite model. First, mixture models have potential to be off-the-shelf tools for practitioners to obtain more flexible posterior approximations in VAEs. Therefore, we make them more accessible by demonstrating how to apply them to four popular architectures. Second, the mixture components cooperate in order to cover the target distribution while trying to maximize their diversity when MISELBO is the objective function. We explain this cooperative behavior by drawing a novel connection between VI and adaptive importance sampling. Finally, we demonstrate the superiority of the Mixture VAEs' learned feature representations on both image and single-cell transcriptome data, and obtain state-of-the-art results among VAE architectures in terms of negative log-likelihood on the MNIST and FashionMNIST datasets. Code available here: \url{https://github.com/Lagergren-Lab/MixtureVAEs}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge