Ramzan Umarov
RNA Secondary Structure Prediction By Learning Unrolled Algorithms
Feb 13, 2020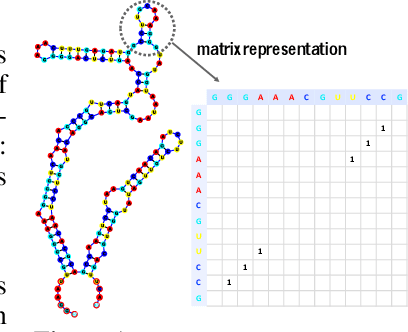
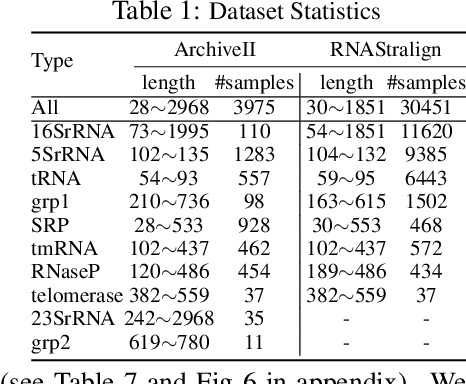
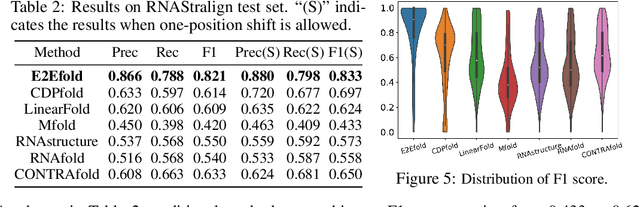
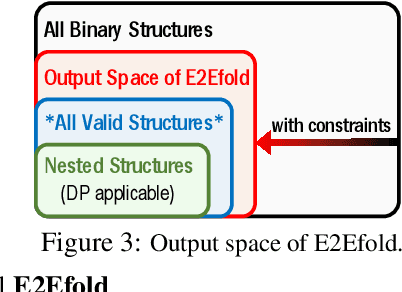
Abstract:In this paper, we propose an end-to-end deep learning model, called E2Efold, for RNA secondary structure prediction which can effectively take into account the inherent constraints in the problem. The key idea of E2Efold is to directly predict the RNA base-pairing matrix, and use an unrolled algorithm for constrained programming as the template for deep architectures to enforce constraints. With comprehensive experiments on benchmark datasets, we demonstrate the superior performance of E2Efold: it predicts significantly better structures compared to previous SOTA (especially for pseudoknotted structures), while being as efficient as the fastest algorithms in terms of inference time.
PromID: human promoter prediction by deep learning
Oct 02, 2018



Abstract:Computational identification of promoters is notoriously difficult as human genes often have unique promoter sequences that provide regulation of transcription and interaction with transcription initiation complex. While there are many attempts to develop computational promoter identification methods, we have no reliable tool to analyze long genomic sequences. In this work we further develop our deep learning approach that was relatively successful to discriminate short promoter and non-promoter sequences. Instead of focusing on the classification accuracy, in this work we predict the exact positions of the TSS inside the genomic sequences testing every possible location. We studied human promoters to find effective regions for discrimination and built corresponding deep learning models. These models use adaptively constructed negative set which iteratively improves the models discriminative ability. The developed promoter identification models significantly outperform the previously developed promoter prediction programs by considerably reducing the number of false positive predictions. The best model we have built has recall 0.76, precision 0.77 and MCC 0.76, while the next best tool FPROM achieved precision 0.48 and MCC 0.60 for the recall of 0.75. Our method is available at http://www.cbrc.kaust.edu.sa/PromID/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge