Rakshith Sathish
SAModified: A Foundation Model-Based Zero-Shot Approach for Refining Noisy Land-Use Land-Cover Maps
Dec 17, 2024



Abstract:Land-use and land cover (LULC) analysis is critical in remote sensing, with wide-ranging applications across diverse fields such as agriculture, utilities, and urban planning. However, automating LULC map generation using machine learning is rendered challenging due to noisy labels. Typically, the ground truths (e.g. ESRI LULC, MapBioMass) have noisy labels that hamper the model's ability to learn to accurately classify the pixels. Further, these erroneous labels can significantly distort the performance metrics of a model, leading to misleading evaluations. Traditionally, the ambiguous labels are rectified using unsupervised algorithms. These algorithms struggle not only with scalability but also with generalization across different geographies. To overcome these challenges, we propose a zero-shot approach using the foundation model, Segment Anything Model (SAM), to automatically delineate different land parcels/regions and leverage them to relabel the unsure pixels by using the local label statistics within each detected region. We achieve a significant reduction in label noise and an improvement in the performance of the downstream segmentation model by $\approx 5\%$ when trained with denoised labels.
Exploiting Richness of Learned Compressed Representation of Images for Semantic Segmentation
Jul 04, 2023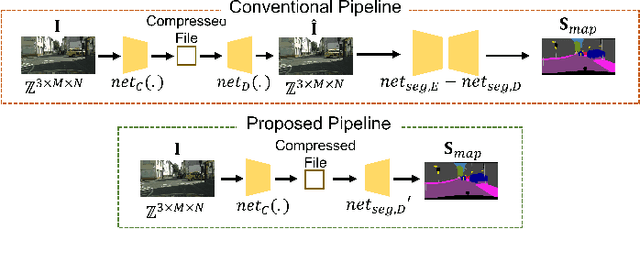
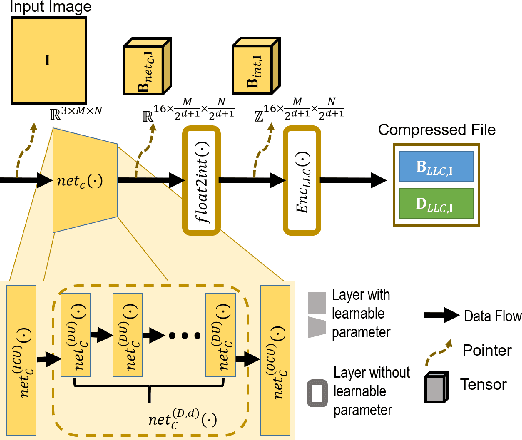
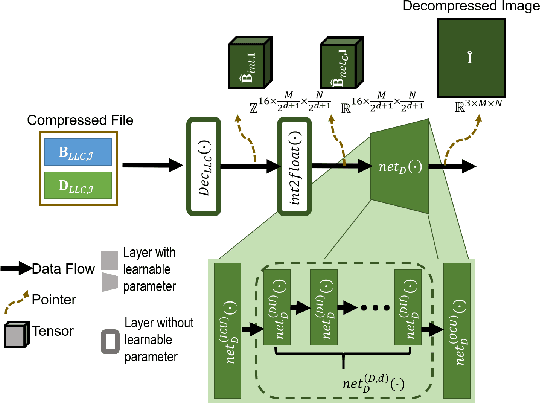
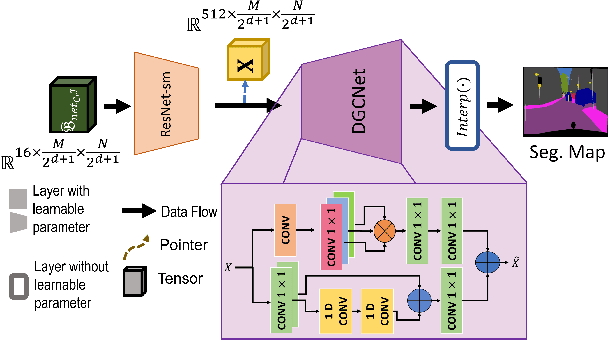
Abstract:Autonomous vehicles and Advanced Driving Assistance Systems (ADAS) have the potential to radically change the way we travel. Many such vehicles currently rely on segmentation and object detection algorithms to detect and track objects around its surrounding. The data collected from the vehicles are often sent to cloud servers to facilitate continual/life-long learning of these algorithms. Considering the bandwidth constraints, the data is compressed before sending it to servers, where it is typically decompressed for training and analysis. In this work, we propose the use of a learning-based compression Codec to reduce the overhead in latency incurred for the decompression operation in the standard pipeline. We demonstrate that the learned compressed representation can also be used to perform tasks like semantic segmentation in addition to decompression to obtain the images. We experimentally validate the proposed pipeline on the Cityscapes dataset, where we achieve a compression factor up to $66 \times$ while preserving the information required to perform segmentation with a dice coefficient of $0.84$ as compared to $0.88$ achieved using decompressed images while reducing the overall compute by $11\%$.
Verifiable and Energy Efficient Medical Image Analysis with Quantised Self-attentive Deep Neural Networks
Sep 30, 2022



Abstract:Convolutional Neural Networks have played a significant role in various medical imaging tasks like classification and segmentation. They provide state-of-the-art performance compared to classical image processing algorithms. However, the major downside of these methods is the high computational complexity, reliance on high-performance hardware like GPUs and the inherent black-box nature of the model. In this paper, we propose quantised stand-alone self-attention based models as an alternative to traditional CNNs. In the proposed class of networks, convolutional layers are replaced with stand-alone self-attention layers, and the network parameters are quantised after training. We experimentally validate the performance of our method on classification and segmentation tasks. We observe a $50-80\%$ reduction in model size, $60-80\%$ lesser number of parameters, $40-85\%$ fewer FLOPs and $65-80\%$ more energy efficiency during inference on CPUs. The code will be available at \href {https://github.com/Rakshith2597/Quantised-Self-Attentive-Deep-Neural-Network}{https://github.com/Rakshith2597/Quantised-Self-Attentive-Deep-Neural-Network}.
Lung Segmentation and Nodule Detection in Computed Tomography Scan using a Convolutional Neural Network Trained Adversarially using Turing Test Loss
Jun 16, 2020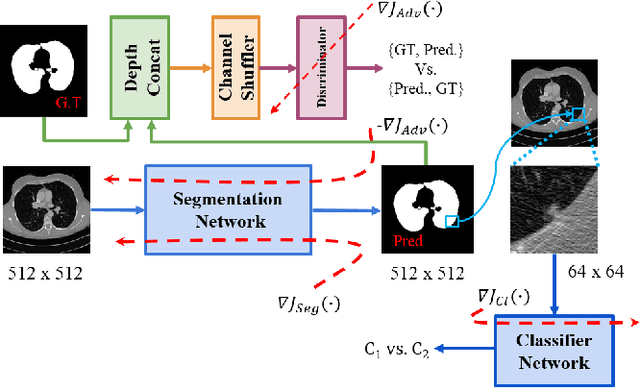
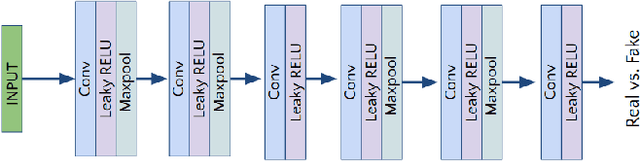
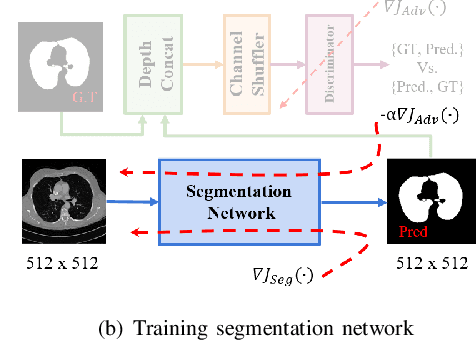
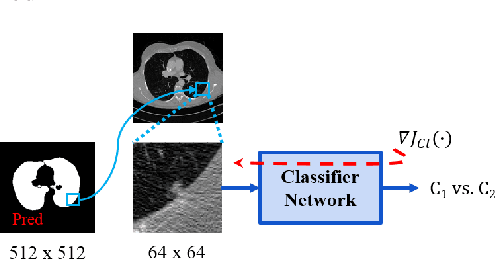
Abstract:Lung cancer is the most common form of cancer found worldwide with a high mortality rate. Early detection of pulmonary nodules by screening with a low-dose computed tomography (CT) scan is crucial for its effective clinical management. Nodules which are symptomatic of malignancy occupy about 0.0125 - 0.025\% of volume in a CT scan of a patient. Manual screening of all slices is a tedious task and presents a high risk of human errors. To tackle this problem we propose a computationally efficient two stage framework. In the first stage, a convolutional neural network (CNN) trained adversarially using Turing test loss segments the lung region. In the second stage, patches sampled from the segmented region are then classified to detect the presence of nodules. The proposed method is experimentally validated on the LUNA16 challenge dataset with a dice coefficient of $0.984\pm0.0007$ for 10-fold cross-validation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge