R. J. G. van Sloun
A Review of Bayesian Uncertainty Quantification in Deep Probabilistic Image Segmentation
Nov 25, 2024Abstract:Advancements in image segmentation play an integral role within the greater scope of Deep Learning-based computer vision. Furthermore, their widespread applicability in critical real-world tasks has given rise to challenges related to the reliability of such algorithms. Hence, uncertainty quantification has been extensively studied within this context, enabling expression of model ignorance (epistemic uncertainty) or data ambiguity (aleatoric uncertainty) to prevent uninformed decision making. Due to the rapid adoption of Convolutional Neural Network (CNN)-based segmentation models in high-stake applications, a substantial body of research has been published on this very topic, causing its swift expansion into a distinct field. This work provides a comprehensive overview of probabilistic segmentation by discussing fundamental concepts in uncertainty that govern advancements in the field as well as the application to various tasks. We identify that quantifying aleatoric and epistemic uncertainty approximates Bayesian inference w.r.t. to either latent variables or model parameters, respectively. Moreover, literature on both uncertainties trace back to four key applications; (1) to quantify statistical inconsistencies in the annotation process due ambiguous images, (2) correlating prediction error with uncertainty, (3) expanding the model hypothesis space for better generalization, and (4) active learning. Then, a discussion follows that includes an overview of utilized datasets for each of the applications and comparison of the available methods. We also highlight challenges related to architectures, uncertainty-based active learning, standardization and benchmarking, and recommendations for future work such as methods based on single forward passes and models that appropriately leverage volumetric data.
Improving Aleatoric Uncertainty Quantification in Multi-Annotated Medical Image Segmentation with Normalizing Flows
Aug 05, 2021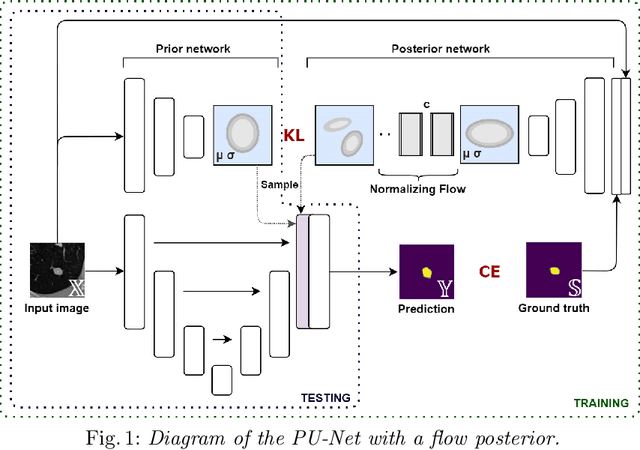

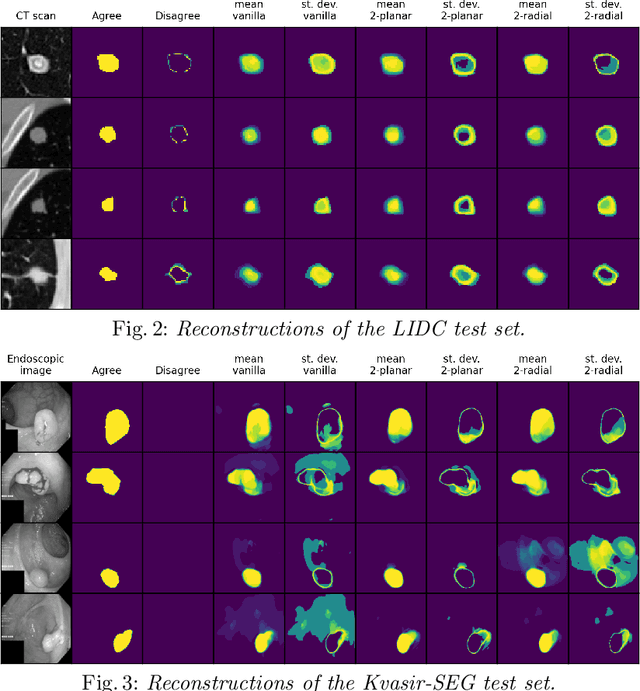
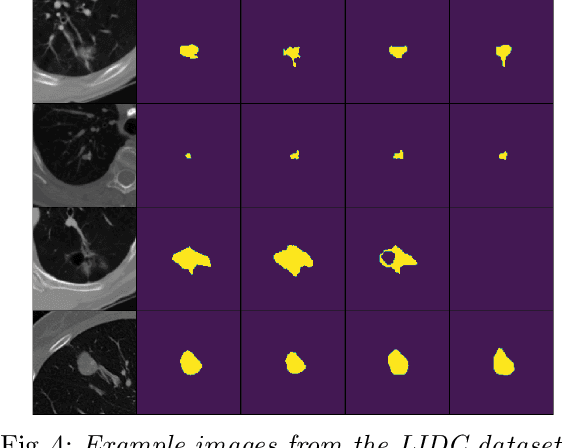
Abstract:Quantifying uncertainty in medical image segmentation applications is essential, as it is often connected to vital decision-making. Compelling attempts have been made in quantifying the uncertainty in image segmentation architectures, e.g. to learn a density segmentation model conditioned on the input image. Typical work in this field restricts these learnt densities to be strictly Gaussian. In this paper, we propose to use a more flexible approach by introducing Normalizing Flows (NFs), which enables the learnt densities to be more complex and facilitate more accurate modeling for uncertainty. We prove this hypothesis by adopting the Probabilistic U-Net and augmenting the posterior density with an NF, allowing it to be more expressive. Our qualitative as well as quantitative (GED and IoU) evaluations on the multi-annotated and single-annotated LIDC-IDRI and Kvasir-SEG segmentation datasets, respectively, show a clear improvement. This is mostly apparent in the quantification of aleatoric uncertainty and the increased predictive performance of up to 14 percent. This result strongly indicates that a more flexible density model should be seriously considered in architectures that attempt to capture segmentation ambiguity through density modeling. The benefit of this improved modeling will increase human confidence in annotation and segmentation, and enable eager adoption of the technology in practice.
Synthetic Elastography using B-mode Ultrasound through a Deep Fully-Convolutional Neural Network
Aug 09, 2019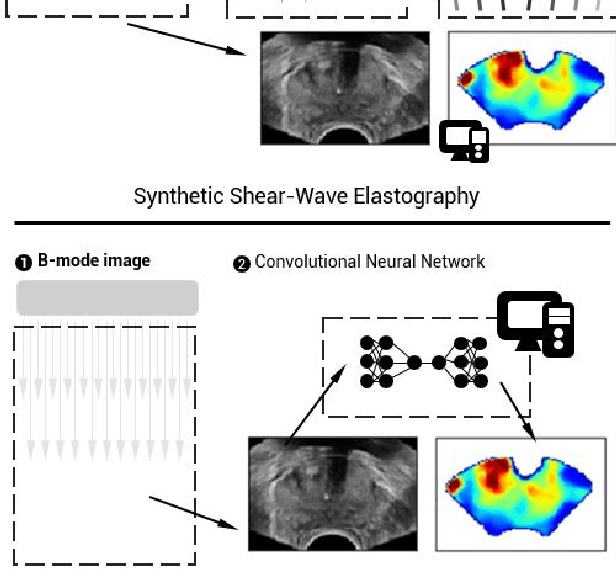
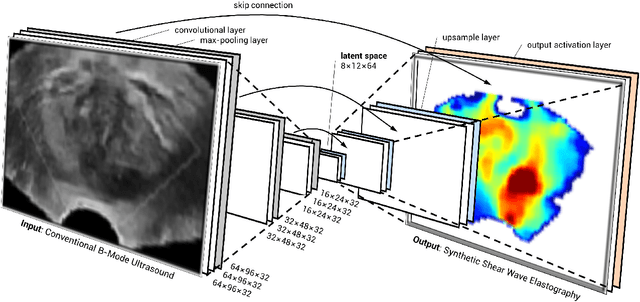
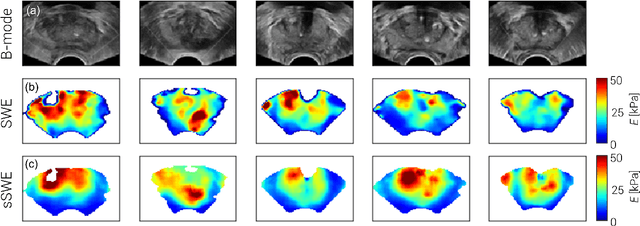
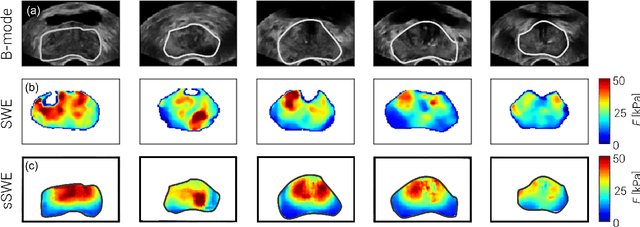
Abstract:Shear-wave elastography (SWE) permits local estimation of tissue elasticity, an important imaging marker in biomedicine. This recently-developed, advanced technique assesses the speed of a laterally-travelling shear wave after an acoustic radiation force "push" to estimate local Young's moduli in an operator-independent fashion. In this work, we show how synthetic SWE (sSWE) images can be generated based on conventional B-mode imaging through deep learning. Using side-by-side-view B-mode/SWE images collected in 50 patients with prostate cancer, we show that sSWE images with a pixel-wise mean absolute error of 4.8 kPa with regard to the original SWE can be generated. Visualization of high-level feature levels through t-Distributed Stochastic Neighbor Embedding reveals a high degree of overlap between data from different scanners. Also qualitatively, sSWE results seem generalisable to single B-mode acquisitions and other scanners. In the future, we envision sSWE as a reliable elasticity-related tissue typing strategy that is solely based on B-mode ultrasound acquisition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge