Improving Aleatoric Uncertainty Quantification in Multi-Annotated Medical Image Segmentation with Normalizing Flows
Paper and Code
Aug 05, 2021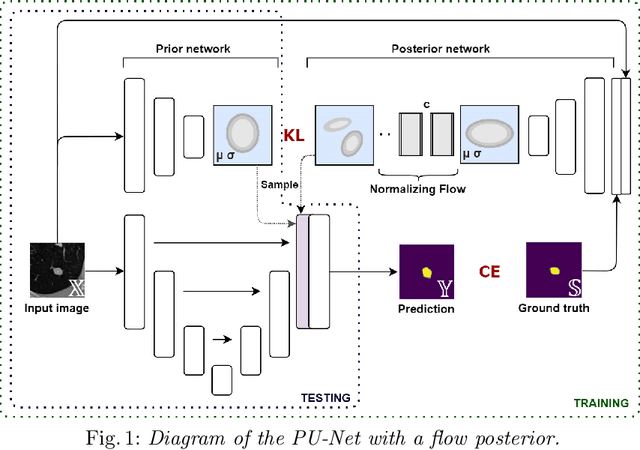

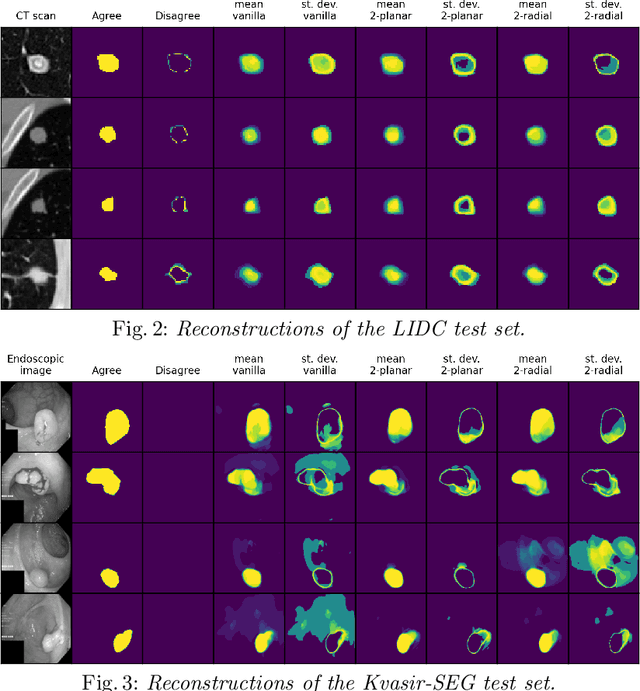
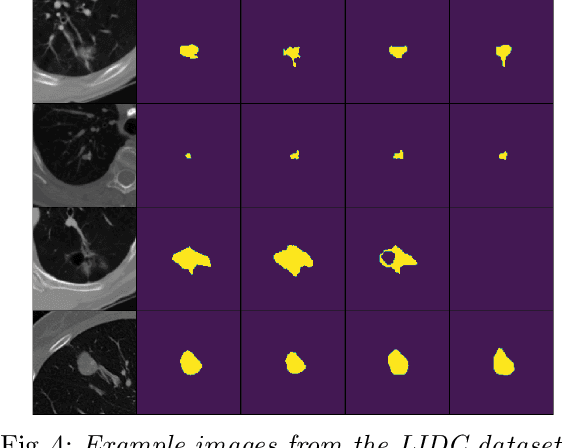
Quantifying uncertainty in medical image segmentation applications is essential, as it is often connected to vital decision-making. Compelling attempts have been made in quantifying the uncertainty in image segmentation architectures, e.g. to learn a density segmentation model conditioned on the input image. Typical work in this field restricts these learnt densities to be strictly Gaussian. In this paper, we propose to use a more flexible approach by introducing Normalizing Flows (NFs), which enables the learnt densities to be more complex and facilitate more accurate modeling for uncertainty. We prove this hypothesis by adopting the Probabilistic U-Net and augmenting the posterior density with an NF, allowing it to be more expressive. Our qualitative as well as quantitative (GED and IoU) evaluations on the multi-annotated and single-annotated LIDC-IDRI and Kvasir-SEG segmentation datasets, respectively, show a clear improvement. This is mostly apparent in the quantification of aleatoric uncertainty and the increased predictive performance of up to 14 percent. This result strongly indicates that a more flexible density model should be seriously considered in architectures that attempt to capture segmentation ambiguity through density modeling. The benefit of this improved modeling will increase human confidence in annotation and segmentation, and enable eager adoption of the technology in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge