Qing En
Cross-model Mutual Learning for Exemplar-based Medical Image Segmentation
Apr 18, 2024



Abstract:Medical image segmentation typically demands extensive dense annotations for model training, which is both time-consuming and skill-intensive. To mitigate this burden, exemplar-based medical image segmentation methods have been introduced to achieve effective training with only one annotated image. In this paper, we introduce a novel Cross-model Mutual learning framework for Exemplar-based Medical image Segmentation (CMEMS), which leverages two models to mutually excavate implicit information from unlabeled data at multiple granularities. CMEMS can eliminate confirmation bias and enable collaborative training to learn complementary information by enforcing consistency at different granularities across models. Concretely, cross-model image perturbation based mutual learning is devised by using weakly perturbed images to generate high-confidence pseudo-labels, supervising predictions of strongly perturbed images across models. This approach enables joint pursuit of prediction consistency at the image granularity. Moreover, cross-model multi-level feature perturbation based mutual learning is designed by letting pseudo-labels supervise predictions from perturbed multi-level features with different resolutions, which can broaden the perturbation space and enhance the robustness of our framework. CMEMS is jointly trained using exemplar data, synthetic data, and unlabeled data in an end-to-end manner. Experimental results on two medical image datasets indicate that the proposed CMEMS outperforms the state-of-the-art segmentation methods with extremely limited supervision.
AKGNet: Attribute Knowledge-Guided Unsupervised Lung-Infected Area Segmentation
Apr 17, 2024Abstract:Lung-infected area segmentation is crucial for assessing the severity of lung diseases. However, existing image-text multi-modal methods typically rely on labour-intensive annotations for model training, posing challenges regarding time and expertise. To address this issue, we propose a novel attribute knowledge-guided framework for unsupervised lung-infected area segmentation (AKGNet), which achieves segmentation solely based on image-text data without any mask annotation. AKGNet facilitates text attribute knowledge learning, attribute-image cross-attention fusion, and high-confidence-based pseudo-label exploration simultaneously. It can learn statistical information and capture spatial correlations between image and text attributes in the embedding space, iteratively refining the mask to enhance segmentation. Specifically, we introduce a text attribute knowledge learning module by extracting attribute knowledge and incorporating it into feature representations, enabling the model to learn statistical information and adapt to different attributes. Moreover, we devise an attribute-image cross-attention module by calculating the correlation between attributes and images in the embedding space to capture spatial dependency information, thus selectively focusing on relevant regions while filtering irrelevant areas. Finally, a self-training mask improvement process is employed by generating pseudo-labels using high-confidence predictions to iteratively enhance the mask and segmentation. Experimental results on a benchmark medical image dataset demonstrate the superior performance of our method compared to state-of-the-art segmentation techniques in unsupervised scenarios.
Efficient Low-Rank GNN Defense Against Structural Attacks
Sep 18, 2023Abstract:Graph Neural Networks (GNNs) have been shown to possess strong representation abilities over graph data. However, GNNs are vulnerable to adversarial attacks, and even minor perturbations to the graph structure can significantly degrade their performance. Existing methods either are ineffective against sophisticated attacks or require the optimization of dense adjacency matrices, which is time-consuming and prone to local minima. To remedy this problem, we propose an Efficient Low-Rank Graph Neural Network (ELR-GNN) defense method, which aims to learn low-rank and sparse graph structures for defending against adversarial attacks, ensuring effective defense with greater efficiency. Specifically, ELR-GNN consists of two modules: a Coarse Low-Rank Estimation Module and a Fine-Grained Estimation Module. The first module adopts the truncated Singular Value Decomposition (SVD) to initialize the low-rank adjacency matrix estimation, which serves as a starting point for optimizing the low-rank matrix. In the second module, the initial estimate is refined by jointly learning a low-rank sparse graph structure with the GNN model. Sparsity is incorporated into the learned low-rank adjacency matrix by pruning weak connections, which can reduce redundant data while maintaining valuable information. As a result, instead of using the dense adjacency matrix directly, ELR-GNN can learn a low-rank and sparse estimate of it in a simple, efficient and easy to optimize manner. The experimental results demonstrate that ELR-GNN outperforms the state-of-the-art GNN defense methods in the literature, in addition to being very efficient and easy to train.
Adaptive Parametric Prototype Learning for Cross-Domain Few-Shot Classification
Sep 04, 2023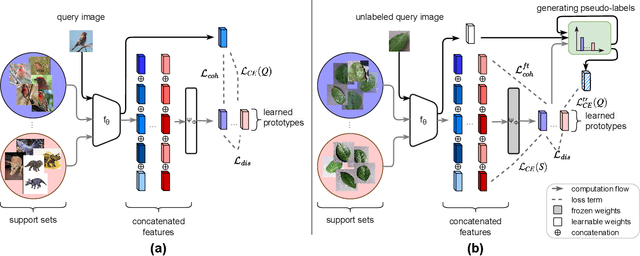
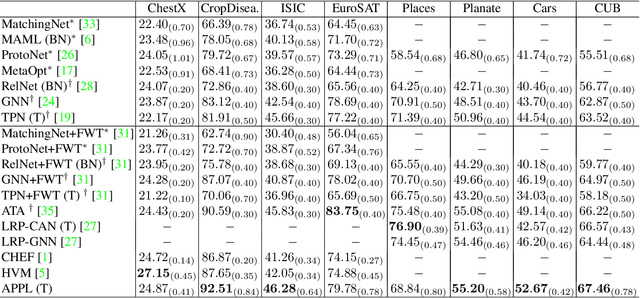
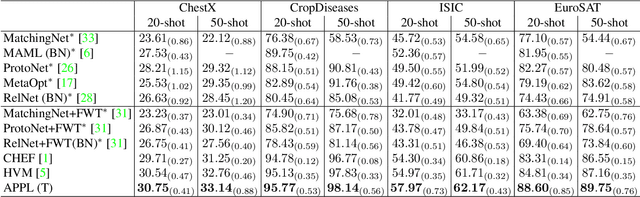
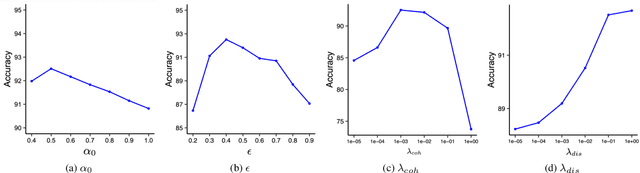
Abstract:Cross-domain few-shot classification induces a much more challenging problem than its in-domain counterpart due to the existence of domain shifts between the training and test tasks. In this paper, we develop a novel Adaptive Parametric Prototype Learning (APPL) method under the meta-learning convention for cross-domain few-shot classification. Different from existing prototypical few-shot methods that use the averages of support instances to calculate the class prototypes, we propose to learn class prototypes from the concatenated features of the support set in a parametric fashion and meta-learn the model by enforcing prototype-based regularization on the query set. In addition, we fine-tune the model in the target domain in a transductive manner using a weighted-moving-average self-training approach on the query instances. We conduct experiments on multiple cross-domain few-shot benchmark datasets. The empirical results demonstrate that APPL yields superior performance than many state-of-the-art cross-domain few-shot learning methods.
Annotation by Clicks: A Point-Supervised Contrastive Variance Method for Medical Semantic Segmentation
Dec 23, 2022Abstract:Medical image segmentation methods typically rely on numerous dense annotated images for model training, which are notoriously expensive and time-consuming to collect. To alleviate this burden, weakly supervised techniques have been exploited to train segmentation models with less expensive annotations. In this paper, we propose a novel point-supervised contrastive variance method (PSCV) for medical image semantic segmentation, which only requires one pixel-point from each organ category to be annotated. The proposed method trains the base segmentation network by using a novel contrastive variance (CV) loss to exploit the unlabeled pixels and a partial cross-entropy loss on the labeled pixels. The CV loss function is designed to exploit the statistical spatial distribution properties of organs in medical images and their variance distribution map representations to enforce discriminative predictions over the unlabeled pixels. Experimental results on two standard medical image datasets demonstrate that the proposed method outperforms the state-of-the-art weakly supervised methods on point-supervised medical image semantic segmentation tasks.
Exemplar Learning for Medical Image Segmentation
Apr 03, 2022



Abstract:Medical image annotation typically requires expert knowledge and hence incurs time-consuming and expensive data annotation costs. To reduce this burden, we propose a novel learning scenario, Exemplar Learning (EL), to explore automated learning processes for medical image segmentation from a single annotated image example. This innovative learning task is particularly suitable for medical image segmentation, where all categories of organs can be presented in one single image for annotation all at once. To address this challenging EL task, we propose an Exemplar Learning-based Synthesis Net (ELSNet) framework for medical image segmentation that enables innovative exemplar-based data synthesis, pixel-prototype based contrastive embedding learning, and pseudo-label based exploitation of the unlabeled data. Specifically, ELSNet introduces two new modules for image segmentation: an exemplar-guided synthesis module, which enriches and diversifies the training set by synthesizing annotated samples from the given exemplar, and a pixel-prototype based contrastive embedding module, which enhances the discriminative capacity of the base segmentation model via contrastive self-supervised learning. Moreover, we deploy a two-stage process for segmentation model training, which exploits the unlabeled data with predicted pseudo segmentation labels. To evaluate this new learning framework, we conduct extensive experiments on several organ segmentation datasets and present an in-depth analysis. The empirical results show that the proposed exemplar learning framework produces effective segmentation results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge