Prakarsh Yadav
Graph Neural Networks Accelerated Molecular Dynamics
Dec 06, 2021

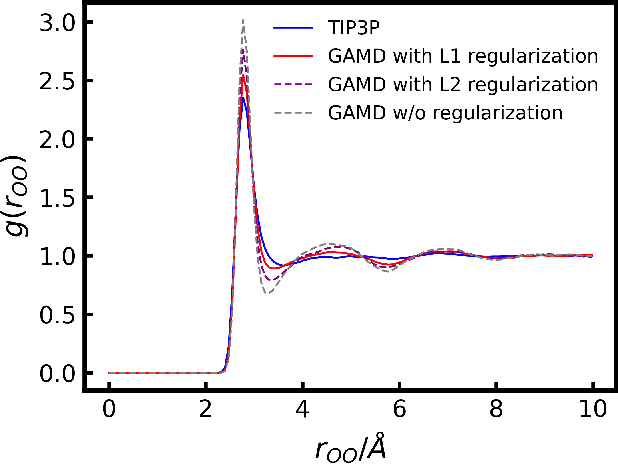
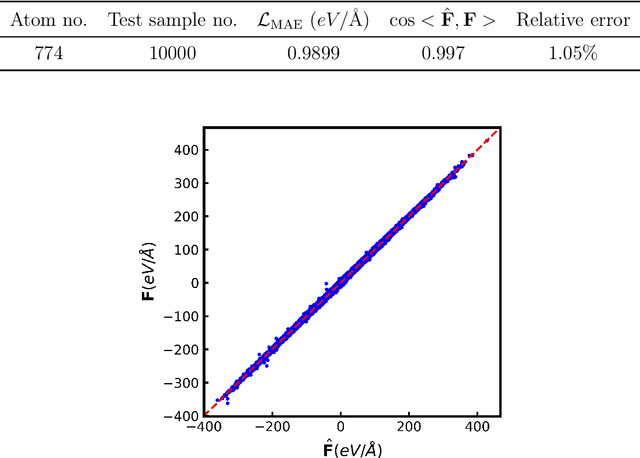
Abstract:Molecular Dynamics (MD) simulation is a powerful tool for understanding the dynamics and structure of matter. Since the resolution of MD is atomic-scale, achieving long time-scale simulations with femtosecond integration is very expensive. In each MD step, numerous redundant computations are performed which can be learnt and avoided. These redundant computations can be surrogated and modeled by a deep learning model like a Graph Neural Network (GNN). In this work, we developed a GNN Accelerated Molecular Dynamics (GAMD) model that achieves fast and accurate force predictions and generates trajectories consistent with the classical MD simulations. Our results show that GAMD can accurately predict the dynamics of two typical molecular systems, Lennard-Jones (LJ) particles and Water (LJ+Electrostatics). GAMD's learning and inference are agnostic to the scale, where it can scale to much larger systems at test time. We also performed a comprehensive benchmark test comparing our implementation of GAMD to production-level MD softwares, where we showed GAMD is competitive with them on the large-scale simulation.
Potential Neutralizing Antibodies Discovered for Novel Corona Virus Using Machine Learning
Mar 18, 2020



Abstract:The fast and untraceable virus mutations take lives of thousands of people before the immune system can produce the inhibitory antibody. Recent outbreak of novel coronavirus infected and killed thousands of people in the world. Rapid methods in finding peptides or antibody sequences that can inhibit the viral epitopes of COVID-19 will save the life of thousands. In this paper, we devised a machine learning (ML) model to predict the possible inhibitory synthetic antibodies for Corona virus. We collected 1933 virus-antibody sequences and their clinical patient neutralization response and trained an ML model to predict the antibody response. Using graph featurization with variety of ML methods, we screened thousands of hypothetical antibody sequences and found 8 stable antibodies that potentially inhibit COVID-19. We combined bioinformatics, structural biology, and Molecular Dynamics (MD) simulations to verify the stability of the candidate antibodies that can inhibit the Corona virus.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge