Pengcheng Zeng
NeuralFLoC: Neural Flow-Based Joint Registration and Clustering of Functional Data
Feb 03, 2026Abstract:Clustering functional data in the presence of phase variation is challenging, as temporal misalignment can obscure intrinsic shape differences and degrade clustering performance. Most existing approaches treat registration and clustering as separate tasks or rely on restrictive parametric assumptions. We present \textbf{NeuralFLoC}, a fully unsupervised, end-to-end deep learning framework for joint functional registration and clustering based on Neural ODE-driven diffeomorphic flows and spectral clustering. The proposed model learns smooth, invertible warping functions and cluster-specific templates simultaneously, effectively disentangling phase and amplitude variation. We establish universal approximation guarantees and asymptotic consistency for the proposed framework. Experiments on functional benchmarks show state-of-the-art performance in both registration and clustering, with robustness to missing data, irregular sampling, and noise, while maintaining scalability. Code is available at https://anonymous.4open.science/r/NeuralFLoC-FEC8.
Semi-parametric Functional Classification via Path Signatures Logistic Regression
Jul 09, 2025Abstract:We propose Path Signatures Logistic Regression (PSLR), a semi-parametric framework for classifying vector-valued functional data with scalar covariates. Classical functional logistic regression models rely on linear assumptions and fixed basis expansions, which limit flexibility and degrade performance under irregular sampling. PSLR overcomes these issues by leveraging truncated path signatures to construct a finite-dimensional, basis-free representation that captures nonlinear and cross-channel dependencies. By embedding trajectories as time-augmented paths, PSLR extracts stable, geometry-aware features that are robust to sampling irregularity without requiring a common time grid, while still preserving subject-specific timing patterns. We establish theoretical guarantees for the existence and consistent estimation of the optimal truncation order, along with non-asymptotic risk bounds. Experiments on synthetic and real-world datasets show that PSLR outperforms traditional functional classifiers in accuracy, robustness, and interpretability, particularly under non-uniform sampling schemes. Our results highlight the practical and theoretical benefits of integrating rough path theory into modern functional data analysis.
DeepFRC: An End-to-End Deep Learning Model for Functional Registration and Classification
Jan 30, 2025



Abstract:Functional data analysis (FDA) is essential for analyzing continuous, high-dimensional data, yet existing methods often decouple functional registration and classification, limiting their efficiency and performance. We present DeepFRC, an end-to-end deep learning framework that unifies these tasks within a single model. Our approach incorporates an alignment module that learns time warping functions via elastic function registration and a learnable basis representation module for dimensionality reduction on aligned data. This integration enhances both alignment accuracy and predictive performance. Theoretical analysis establishes that DeepFRC achieves low misalignment and generalization error, while simulations elucidate the progression of registration, reconstruction, and classification during training. Experiments on real-world datasets demonstrate that DeepFRC consistently outperforms state-of-the-art methods, particularly in addressing complex registration challenges. Code is available at: https://github.com/Drivergo-93589/DeepFRC.
Learnable Blur Kernel for Single-Image Defocus Deblurring in the Wild
Nov 25, 2022Abstract:Recent research showed that the dual-pixel sensor has made great progress in defocus map estimation and image defocus deblurring. However, extracting real-time dual-pixel views is troublesome and complex in algorithm deployment. Moreover, the deblurred image generated by the defocus deblurring network lacks high-frequency details, which is unsatisfactory in human perception. To overcome this issue, we propose a novel defocus deblurring method that uses the guidance of the defocus map to implement image deblurring. The proposed method consists of a learnable blur kernel to estimate the defocus map, which is an unsupervised method, and a single-image defocus deblurring generative adversarial network (DefocusGAN) for the first time. The proposed network can learn the deblurring of different regions and recover realistic details. We propose a defocus adversarial loss to guide this training process. Competitive experimental results confirm that with a learnable blur kernel, the generated defocus map can achieve results comparable to supervised methods. In the single-image defocus deblurring task, the proposed method achieves state-of-the-art results, especially significant improvements in perceptual quality, where PSNR reaches 25.56 dB and LPIPS reaches 0.111.
scICML: Information-theoretic Co-clustering-based Multi-view Learning for the Integrative Analysis of Single-cell Multi-omics data
May 19, 2022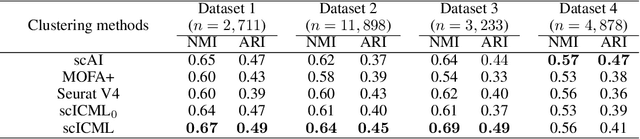


Abstract:Modern high-throughput sequencing technologies have enabled us to profile multiple molecular modalities from the same single cell, providing unprecedented opportunities to assay celluar heterogeneity from multiple biological layers. However, the datasets generated from these technologies tend to have high level of noise and are highly sparse, bringing challenges to data analysis. In this paper, we develop a novel information-theoretic co-clustering-based multi-view learning (scICML) method for multi-omics single-cell data integration. scICML utilizes co-clusterings to aggregate similar features for each view of data and uncover the common clustering pattern for cells. In addition, scICML automatically matches the clusters of the linked features across different data types for considering the biological dependency structure across different types of genomic features. Our experiments on four real-world datasets demonstrate that scICML improves the overall clustering performance and provides biological insights into the data analysis of peripheral blood mononuclear cells.
Elastic Coupled Co-clustering for Single-Cell Genomic Data
Mar 29, 2020


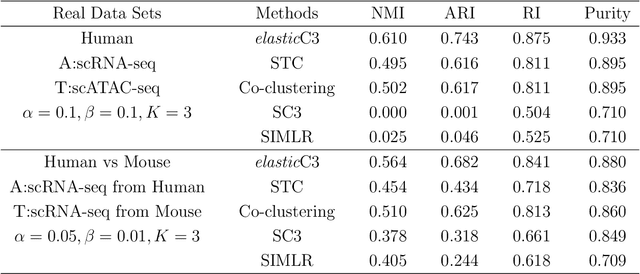
Abstract:The recent advances in single-cell technologies have enabled us to profile genomic features at unprecedented resolution and data sets from multiple domains are available, including data sets that profile different types of genomic features and data sets that profile the same type of genomic features across different species. These data sets typically have different powers in identifying the unknown cell types through clustering, and data integration can potentially lead to a better performance of clustering algorithms. In this work, we formulate the problem in an unsupervised transfer learning framework, which utilizes knowledge learned from auxiliary data set to improve the clustering performance of target data set. The degree of shared information among the target and auxiliary data sets can vary, and their distributions can also be different. To address these challenges, we propose an elastic coupled co-clustering based transfer learning algorithm, by elastically propagating clustering knowledge obtained from the auxiliary data set to the target data set. Implementation on single-cell genomic data sets shows that our algorithm greatly improves clustering performance over the traditional learning algorithms. The source code and data sets are available at https://github.com/cuhklinlab/elasticC3.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge