Zhixiang Lin
scICML: Information-theoretic Co-clustering-based Multi-view Learning for the Integrative Analysis of Single-cell Multi-omics data
May 19, 2022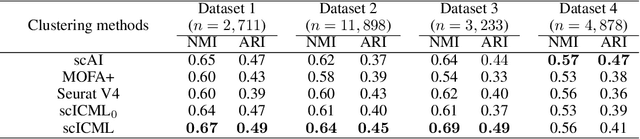


Abstract:Modern high-throughput sequencing technologies have enabled us to profile multiple molecular modalities from the same single cell, providing unprecedented opportunities to assay celluar heterogeneity from multiple biological layers. However, the datasets generated from these technologies tend to have high level of noise and are highly sparse, bringing challenges to data analysis. In this paper, we develop a novel information-theoretic co-clustering-based multi-view learning (scICML) method for multi-omics single-cell data integration. scICML utilizes co-clusterings to aggregate similar features for each view of data and uncover the common clustering pattern for cells. In addition, scICML automatically matches the clusters of the linked features across different data types for considering the biological dependency structure across different types of genomic features. Our experiments on four real-world datasets demonstrate that scICML improves the overall clustering performance and provides biological insights into the data analysis of peripheral blood mononuclear cells.
Elastic Coupled Co-clustering for Single-Cell Genomic Data
Mar 29, 2020


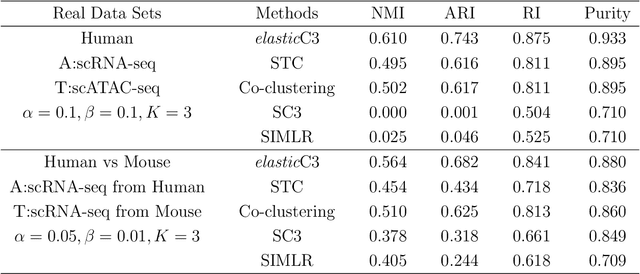
Abstract:The recent advances in single-cell technologies have enabled us to profile genomic features at unprecedented resolution and data sets from multiple domains are available, including data sets that profile different types of genomic features and data sets that profile the same type of genomic features across different species. These data sets typically have different powers in identifying the unknown cell types through clustering, and data integration can potentially lead to a better performance of clustering algorithms. In this work, we formulate the problem in an unsupervised transfer learning framework, which utilizes knowledge learned from auxiliary data set to improve the clustering performance of target data set. The degree of shared information among the target and auxiliary data sets can vary, and their distributions can also be different. To address these challenges, we propose an elastic coupled co-clustering based transfer learning algorithm, by elastically propagating clustering knowledge obtained from the auxiliary data set to the target data set. Implementation on single-cell genomic data sets shows that our algorithm greatly improves clustering performance over the traditional learning algorithms. The source code and data sets are available at https://github.com/cuhklinlab/elasticC3.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge