Paule-J Toussaint
McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University, Montreal, QC, Canada
Confidence-Guided Unsupervised Domain Adaptation for Cerebellum Segmentation
Jun 14, 2022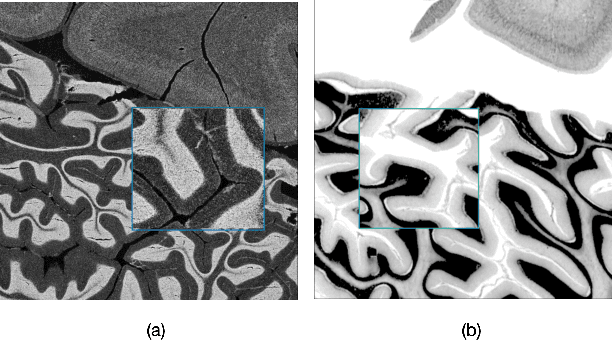
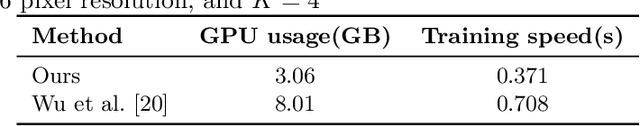
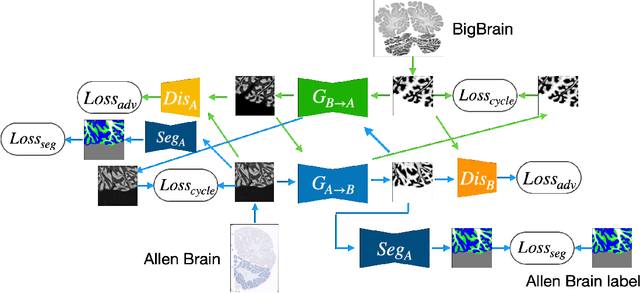
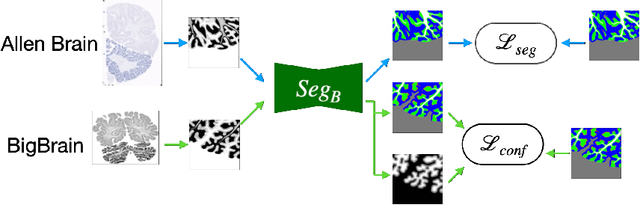
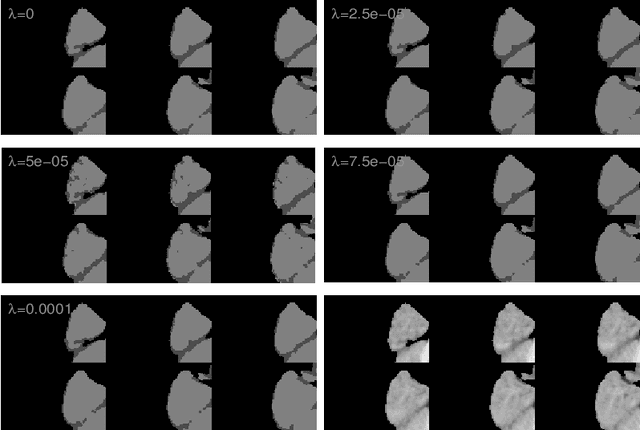
Abstract:The lack of a comprehensive high-resolution atlas of the cerebellum has hampered studies of cerebellar involvement in normal brain function and disease. A good representation of the tightly foliated aspect of the cerebellar cortex is difficult to achieve because of the highly convoluted surface and the time it would take for manual delineation. The quality of manual segmentation is influenced by human expert judgment, and automatic labelling is constrained by the limited robustness of existing segmentation algorithms. The 20umisotropic BigBrain dataset provides an unprecedented high resolution framework for semantic segmentation compared to the 1000um(1mm) resolution afforded by magnetic resonance imaging. To dispense with the manual annotation requirement, we propose to train a model to adaptively transfer the annotation from the cerebellum on the Allen Brain Human Brain Atlas to the BigBrain in an unsupervised manner, taking into account the different staining and spacing between sections. The distinct visual discrepancy between the Allen Brain and BigBrain prevents existing approaches to provide meaningful segmentation masks, and artifacts caused by sectioning and histological slice preparation in the BigBrain data pose an extra challenge. To address these problems, we propose a two-stage framework where we first transfer the Allen Brain cerebellum to a space sharing visual similarity with the BigBrain. We then introduce a self-training strategy with a confidence map to guide the model learning from the noisy pseudo labels iteratively. Qualitative results validate the effectiveness of our approach, and quantitative experiments reveal that our method can achieve over 2.6% loss reduction compared with other approaches.
Local semi-supervised approach to brain tissue classification in child brain MRI
May 20, 2020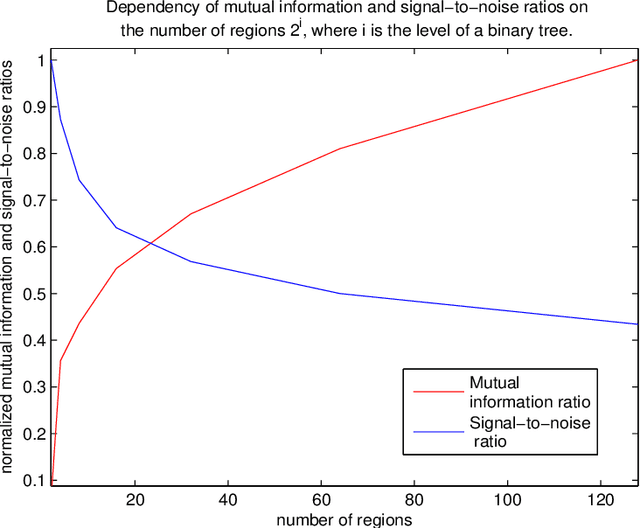
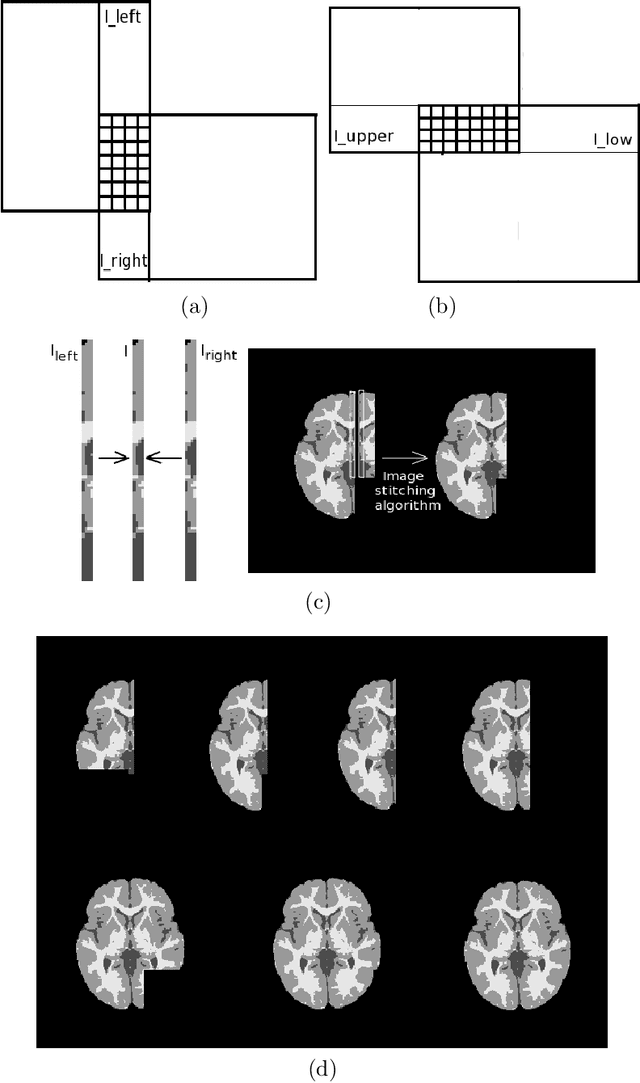
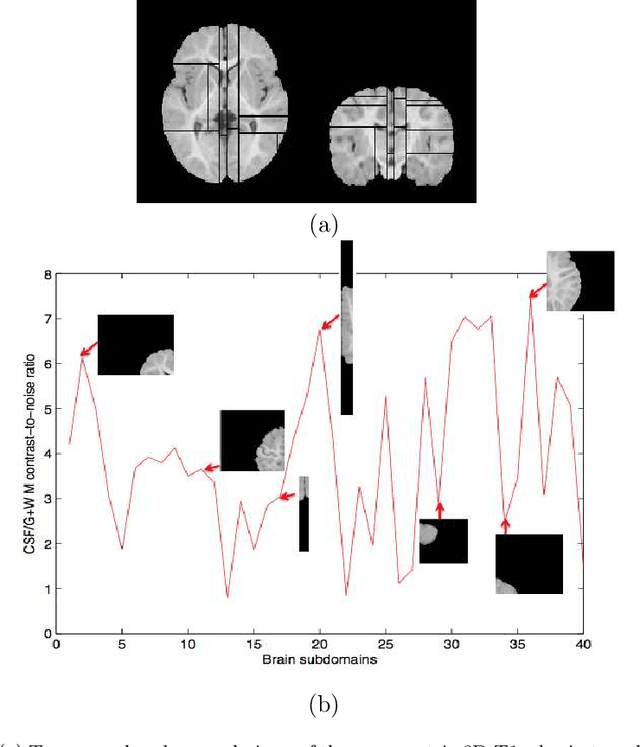
Abstract:Most segmentation methods in child brain MRI are supervised and are based on global intensity distributions of major brain structures. The successful implementation of a supervised approach depends on availability of an age-appropriate probabilistic brain atlas. For the study of early normal brain development, the construction of such a brain atlas remains a significant challenge. Moreover, using global intensity statistics leads to inaccurate detection of major brain tissue classes due to substantial intensity variations of MR signal within the constituent parts of early developing brain. In order to overcome these methodological limitations we develop a local, semi-supervised framework. It is based on Kernel Fisher Discriminant Analysis (KFDA) for pattern recognition, combined with an objective structural similarity index (SSIM) for perceptual image quality assessment. The proposed method performs optimal brain partitioning into subdomains having different average intensity values followed by SSIM-guided computation of separating surfaces between the constituent brain parts. The classified image subdomains are then stitched slice by slice via simulated annealing to form a global image of the classified brain. In this paper, we consider classification into major tissue classes (white matter and grey matter) and the cerebrospinal fluid and illustrate the proposed framework on examples of brain templates for ages 8 to 11 months and ages 44 to 60 months. We show that our method improves detection of the tissue classes by its comparison to state-of-the-art classification techniques known as Partial Volume Estimation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge