Or Dinari
CPU- and GPU-based Distributed Sampling in Dirichlet Process Mixtures for Large-scale Analysis
Apr 19, 2022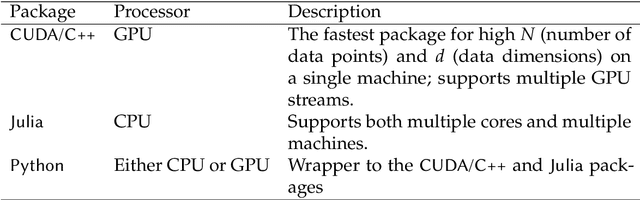
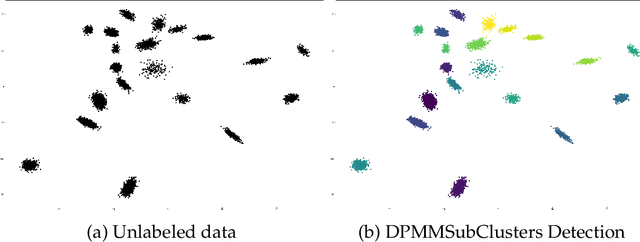

Abstract:In the realm of unsupervised learning, Bayesian nonparametric mixture models, exemplified by the Dirichlet Process Mixture Model (DPMM), provide a principled approach for adapting the complexity of the model to the data. Such models are particularly useful in clustering tasks where the number of clusters is unknown. Despite their potential and mathematical elegance, however, DPMMs have yet to become a mainstream tool widely adopted by practitioners. This is arguably due to a misconception that these models scale poorly as well as the lack of high-performance (and user-friendly) software tools that can handle large datasets efficiently. In this paper we bridge this practical gap by proposing a new, easy-to-use, statistical software package for scalable DPMM inference. More concretely, we provide efficient and easily-modifiable implementations for high-performance distributed sampling-based inference in DPMMs where the user is free to choose between either a multiple-machine, multiple-core, CPU implementation (written in Julia) and a multiple-stream GPU implementation (written in CUDA/C++). Both the CPU and GPU implementations come with a common (and optional) python wrapper, providing the user with a single point of entry with the same interface. On the algorithmic side, our implementations leverage a leading DPMM sampler from (Chang and Fisher III, 2013). While Chang and Fisher III's implementation (written in MATLAB/C++) used only CPU and was designed for a single multi-core machine, the packages we proposed here distribute the computations efficiently across either multiple multi-core machines or across mutiple GPU streams. This leads to speedups, alleviates memory and storage limitations, and lets us fit DPMMs to significantly larger datasets and of higher dimensionality than was possible previously by either (Chang and Fisher III, 2013) or other DPMM methods.
Common Failure Modes of Subcluster-based Sampling in Dirichlet Process Gaussian Mixture Models -- and a Deep-learning Solution
Mar 25, 2022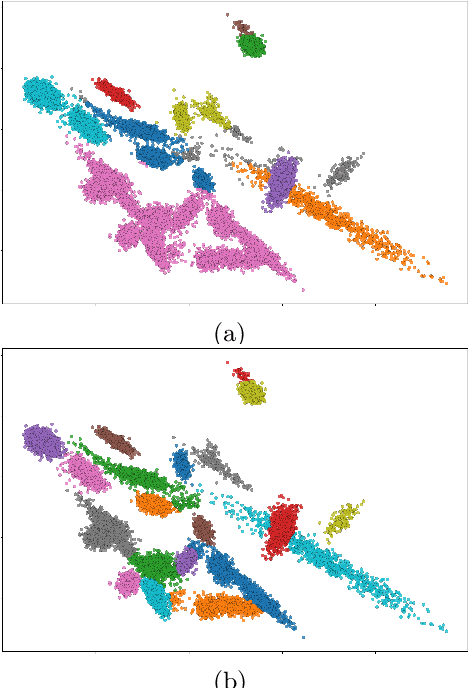
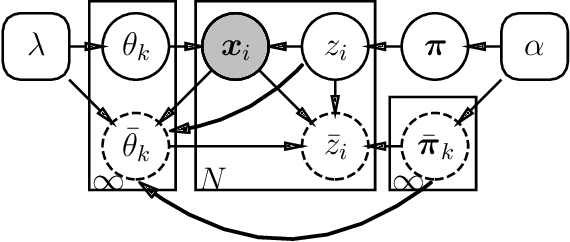
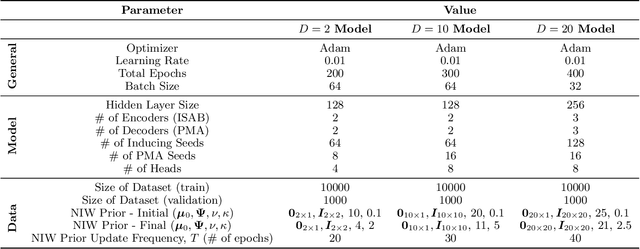
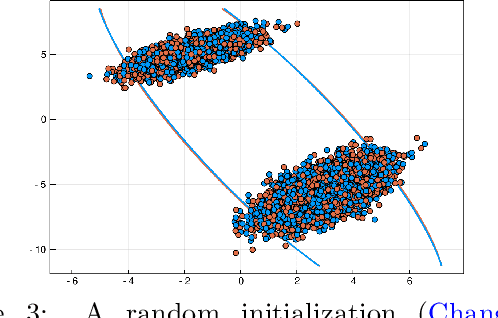
Abstract:The Dirichlet Process Gaussian Mixture Model (DPGMM) is often used to cluster data when the number of clusters is unknown. One main DPGMM inference paradigm relies on sampling. Here we consider a known state-of-art sampler (proposed by Chang and Fisher III (2013) and improved by Dinari et al. (2019)), analyze its failure modes, and show how to improve it, often drastically. Concretely, in that sampler, whenever a new cluster is formed it is augmented with two subclusters whose labels are initialized at random. Upon their evolution, the subclusters serve to propose a split of the parent cluster. We show that the random initialization is often problematic and hurts the otherwise-effective sampler. Specifically, we demonstrate that this initialization tends to lead to poor split proposals and/or too many iterations before a desired split is accepted. This slows convergence and can damage the clustering. As a remedy, we propose two drop-in-replacement options for the subcluster-initialization subroutine. The first is an intuitive heuristic while the second is based on deep learning. We show that the proposed approach yields better splits, which in turn translate to substantial improvements in performance, results, and stability.
Sampling in Dirichlet Process Mixture Models for Clustering Streaming Data
Feb 27, 2022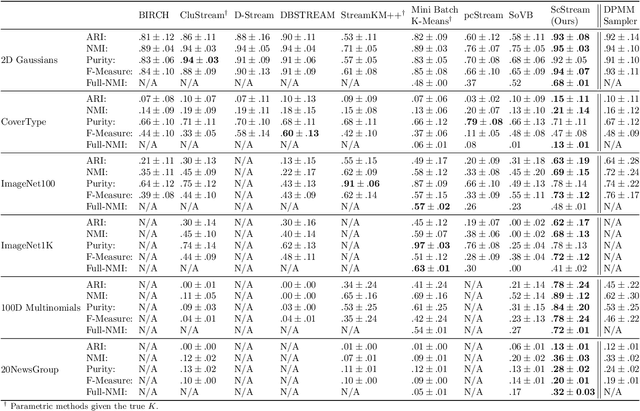

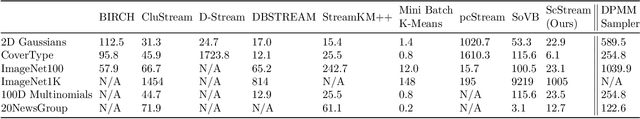

Abstract:Practical tools for clustering streaming data must be fast enough to handle the arrival rate of the observations. Typically, they also must adapt on the fly to possible lack of stationarity; i.e., the data statistics may be time-dependent due to various forms of drifts, changes in the number of clusters, etc. The Dirichlet Process Mixture Model (DPMM), whose Bayesian nonparametric nature allows it to adapt its complexity to the data, seems a natural choice for the streaming-data case. In its classical formulation, however, the DPMM cannot capture common types of drifts in the data statistics. Moreover, and regardless of that limitation, existing methods for online DPMM inference are too slow to handle rapid data streams. In this work we propose adapting both the DPMM and a known DPMM sampling-based non-streaming inference method for streaming-data clustering. We demonstrate the utility of the proposed method on several challenging settings, where it obtains state-of-the-art results while being on par with other methods in terms of speed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge