Noam Harel
Deep Cerebellar Nuclei Segmentation via Semi-Supervised Deep Context-Aware Learning from 7T Diffusion MRI
May 12, 2020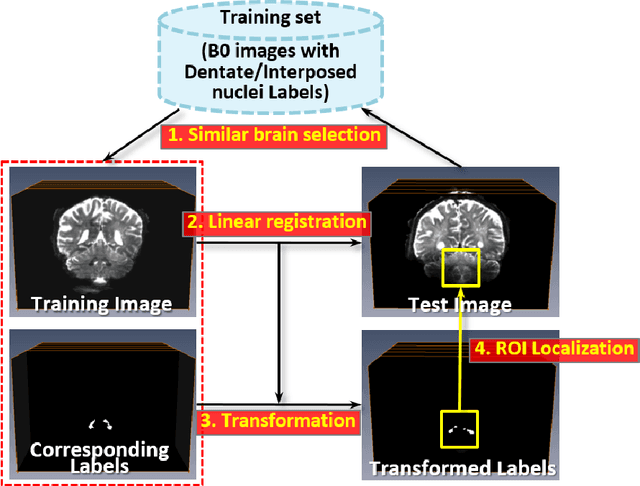

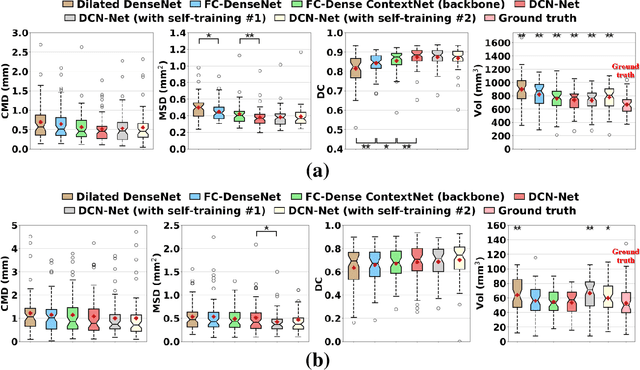
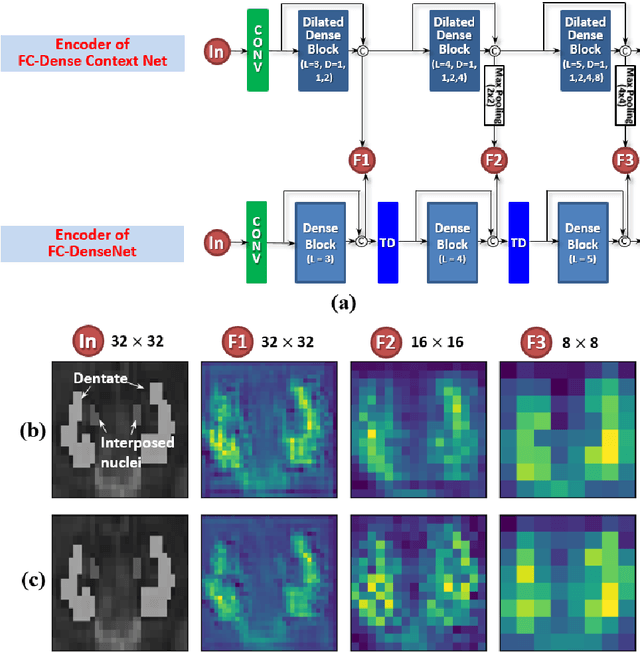
Abstract:Deep cerebellar nuclei are a key structure of the cerebellum that are involved in processing motor and sensory information. It is thus a crucial step to accurately segment deep cerebellar nuclei for the understanding of the cerebellum system and its utility in deep brain stimulation treatment. However, it is challenging to clearly visualize such small nuclei under standard clinical magnetic resonance imaging (MRI) protocols and therefore precise segmentation is not feasible. Recent advances in 7 Tesla (T) MRI technology and great potential of deep neural networks facilitate automatic patient-specific segmentation. In this paper, we propose a novel deep learning framework (referred to as DCN-Net) for fast, accurate, and robust patient-specific segmentation of deep cerebellar dentate and interposed nuclei on 7T diffusion MRI. DCN-Net effectively encodes contextual information on the patch images without consecutive pooling operations and adding complexity via proposed dilated dense blocks. During the end-to-end training, label probabilities of dentate and interposed nuclei are independently learned with a hybrid loss, handling highly imbalanced data. Finally, we utilize self-training strategies to cope with the problem of limited labeled data. To this end, auxiliary dentate and interposed nuclei labels are created on unlabeled data by using DCN-Net trained on manual labels. We validate the proposed framework using 7T B0 MRIs from 60 subjects. Experimental results demonstrate that DCN-Net provides better segmentation than atlas-based deep cerebellar nuclei segmentation tools and other state-of-the-art deep neural networks in terms of accuracy and consistency. We further prove the effectiveness of the proposed components within DCN-Net in dentate and interposed nuclei segmentation.
Continuous Dice Coefficient: a Method for Evaluating Probabilistic Segmentations
Jun 26, 2019


Abstract:Objective: Overlapping measures are often utilized to quantify the similarity between two binary regions. However, modern segmentation algorithms output a probability or confidence map with continuous values in the zero-to-one interval. Moreover, these binary overlapping measures are biased to structure size. Addressing these challenges is the objective of this work. Methods: We extend the definition of the classical Dice coefficient (DC) overlap to facilitate the direct comparison of a ground truth binary image with a probabilistic map. We call the extended method continuous Dice coefficient (cDC) and show that 1) cDC is less or equal to 1 and cDC = 1 if-and-only-if the structures overlap is complete, and, 2) cDC is monotonically decreasing with the amount of overlap. We compare the classical DC and the cDC in a simulation of partial volume effects that incorporates segmentations of common targets for deep-brainstimulation. Lastly, we investigate the cDC for an automatic segmentation of the subthalamic-nucleus. Results: Partial volume effect simulation on thalamus (large structure) resulted with DC and cDC averages (SD) of 0.98 (0.006) and 0.99 (0.001), respectively. For subthalamic-nucleus (small structure) DC and cDC were 0.86 (0.025) and 0.97 (0.006), respectively. The DC and cDC for automatic STN segmentation were 0.66 and 0.80, respectively. Conclusion: The cDC is well defined for probabilistic segmentation, less biased to structure size and more robust to partial volume effects in comparison to DC. Significance: The proposed method facilitates a better evaluation of segmentation algorithms. As a better measurement tool, it opens the door for the development of better segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge