Nicolas J-B. Brunel
Efficient Precision Control in Object Detection Models for Enhanced and Reliable Ovarian Follicle Counting
Jan 23, 2025Abstract:Image analysis is a key tool for describing the detailed mechanisms of folliculogenesis, such as evaluating the quantity of mouse Primordial ovarian Follicles (PMF) in the ovarian reserve. The development of high-resolution virtual slide scanners offers the possibility of quantifying, robustifying and accelerating the histopathological procedure. A major challenge for machine learning is to control the precision of predictions while enabling a high recall, in order to provide reproducibility. We use a multiple testing procedure that gives an overperforming way to solve the standard Precision-Recall trade-off that gives probabilistic guarantees on the precision. In addition, we significantly improve the overall performance of the models (increase of F1-score) by selecting the decision threshold using contextual biological information or using an auxiliary model. As it is model-agnostic, this contextual selection procedure paves the way to the development of a strategy that can improve the performance of any model without the need of retraining it.
Accurate and robust Shapley Values for explaining predictions and focusing on local important variables
Jun 07, 2021



Abstract:Although Shapley Values (SV) are widely used in explainable AI, they can be poorly understood and estimated, which implies that their analysis may lead to spurious inferences and explanations. As a starting point, we remind an invariance principle for SV and derive the correct approach for computing the SV of categorical variables that are particularly sensitive to the encoding used. In the case of tree-based models, we introduce two estimators of Shapley Values that exploit efficiently the tree structure and are more accurate than state-of-the-art methods. For interpreting additive explanations, we recommend to filter the non-influential variables and to compute the Shapley Values only for groups of influential variables. For this purpose, we use the concept of "Same Decision Probability" (SDP) that evaluates the robustness of a prediction when some variables are missing. This prior selection procedure produces sparse additive explanations easier to visualize and analyse. Simulations and comparisons are performed with state-of-the-art algorithm, and show the practical gain of our approach.
The Shapley Value of coalition of variables provides better explanations
Mar 25, 2021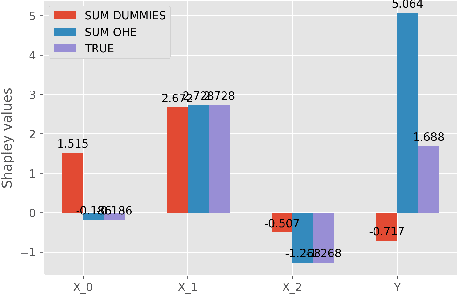
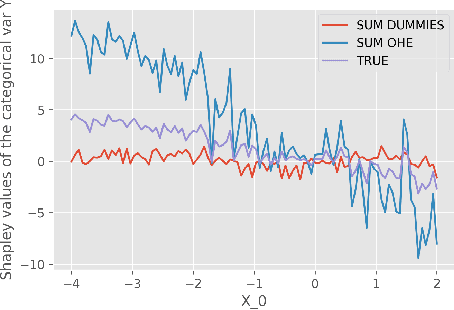
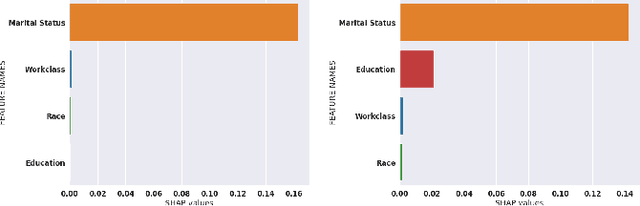
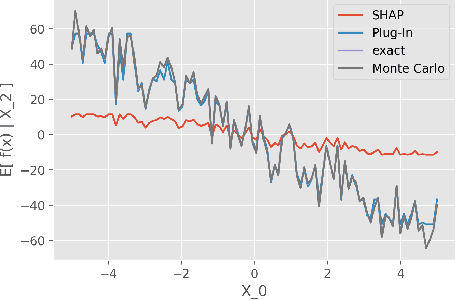
Abstract:While Shapley Values (SV) are one of the gold standard for interpreting machine learning models, we show that they are still poorly understood, in particular in the presence of categorical variables or of variables of low importance. For instance, we show that the popular practice that consists in summing the SV of dummy variables is false as it provides wrong estimates of all the SV in the model and implies spurious interpretations. Based on the identification of null and active coalitions, and a coalitional version of the SV, we provide a correct computation and inference of important variables. Moreover, a Python library (All the experiments and simulations can be reproduced with the publicly available library Active Coalition of Variables, https://www.github.com/salimamoukou/acv00) that computes reliably conditional expectations and SV for tree-based models, is implemented and compared with state-of-the-art algorithms on toy models and real data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge