Nico Schiavone
Reinforcement Learning with Generative Models for Compact Support Sets
Apr 25, 2024Abstract:Foundation models contain a wealth of information from their vast number of training samples. However, most prior arts fail to extract this information in a precise and efficient way for small sample sizes. In this work, we propose a framework utilizing reinforcement learning as a control for foundation models, allowing for the granular generation of small, focused synthetic support sets to augment the performance of neural network models on real data classification tasks. We first allow a reinforcement learning agent access to a novel context based dictionary; the agent then uses this dictionary with a novel prompt structure to form and optimize prompts as inputs to generative models, receiving feedback based on a reward function combining the change in validation accuracy and entropy. A support set is formed this way over several exploration steps. Our framework produced excellent results, increasing classification accuracy by significant margins for no additional labelling or data cost.
MyriadAL: Active Few Shot Learning for Histopathology
Oct 24, 2023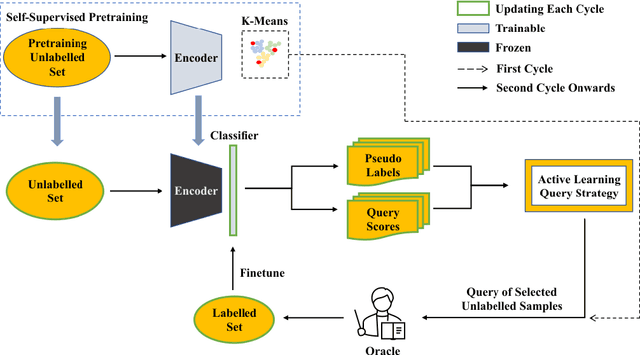

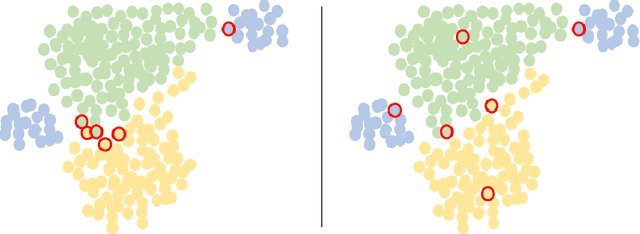
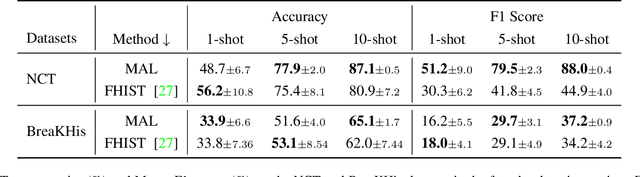
Abstract:Active Learning (AL) and Few Shot Learning (FSL) are two label-efficient methods which have achieved excellent results recently. However, most prior arts in both learning paradigms fail to explore the wealth of the vast unlabelled data. In this study, we address this issue in the scenario where the annotation budget is very limited, yet a large amount of unlabelled data for the target task is available. We frame this work in the context of histopathology where labelling is prohibitively expensive. To this end, we introduce an active few shot learning framework, Myriad Active Learning (MAL), including a contrastive-learning encoder, pseudo-label generation, and novel query sample selection in the loop. Specifically, we propose to massage unlabelled data in a self-supervised manner, where the obtained data representations and clustering knowledge form the basis to activate the AL loop. With feedback from the oracle in each AL cycle, the pseudo-labels of the unlabelled data are refined by optimizing a shallow task-specific net on top of the encoder. These updated pseudo-labels serve to inform and improve the active learning query selection process. Furthermore, we introduce a novel recipe to combine existing uncertainty measures and utilize the entire uncertainty list to reduce sample redundancy in AL. Extensive experiments on two public histopathology datasets show that MAL has superior test accuracy, macro F1-score, and label efficiency compared to prior works, and can achieve a comparable test accuracy to a fully supervised algorithm while labelling only 5% of the dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge