Neeraj Dhungel
Automated Detection of Individual Micro-calcifications from Mammograms using a Multi-stage Cascade Approach
Oct 07, 2016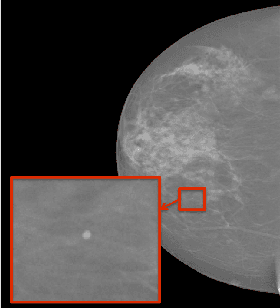
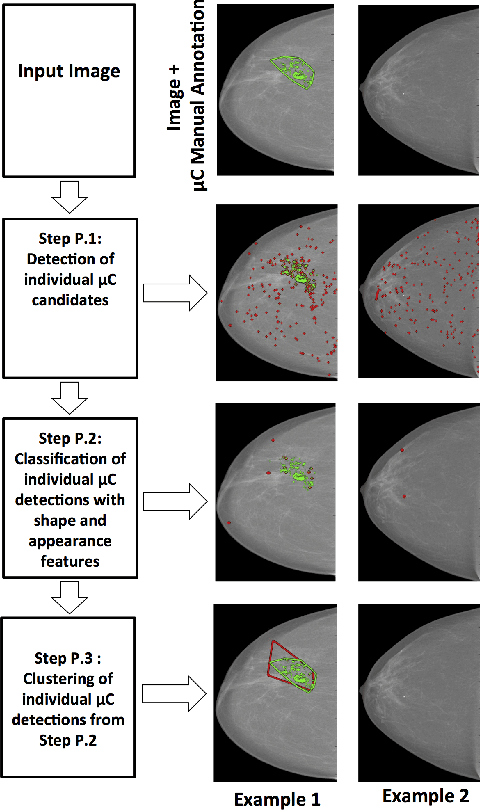
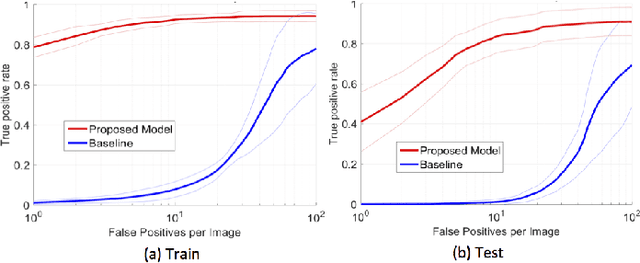
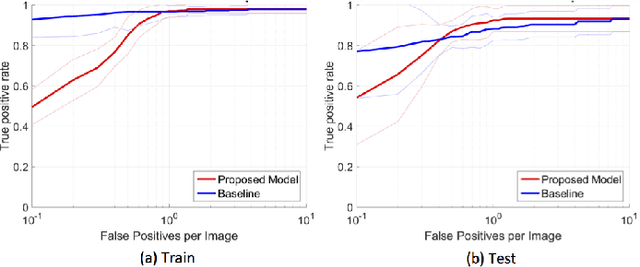
Abstract:In mammography, the efficacy of computer-aided detection methods depends, in part, on the robust localisation of micro-calcifications ($\mu$C). Currently, the most effective methods are based on three steps: 1) detection of individual $\mu$C candidates, 2) clustering of individual $\mu$C candidates, and 3) classification of $\mu$C clusters. Where the second step is motivated both to reduce the number of false positive detections from the first step and on the evidence that malignancy depends on a relatively large number of $\mu$C detections within a certain area. In this paper, we propose a novel approach to $\mu$C detection, consisting of the detection \emph{and} classification of individual $\mu$C candidates, using shape and appearance features, using a cascade of boosting classifiers. The final step in our approach then clusters the remaining individual $\mu$C candidates. The main advantage of this approach lies in its ability to reject a significant number of false positive $\mu$C candidates compared to previously proposed methods. Specifically, on the INbreast dataset, we show that our approach has a true positive rate (TPR) for individual $\mu$Cs of 40\% at one false positive per image (FPI) and a TPR of 80\% at 10 FPI. These results are significantly more accurate than the current state of the art, which has a TPR of less than 1\% at one FPI and a TPR of 10\% at 10 FPI. Our results are competitive with the state of the art at the subsequent stage of detecting clusters of $\mu$Cs.
Deep Structured learning for mass segmentation from Mammograms
Dec 05, 2014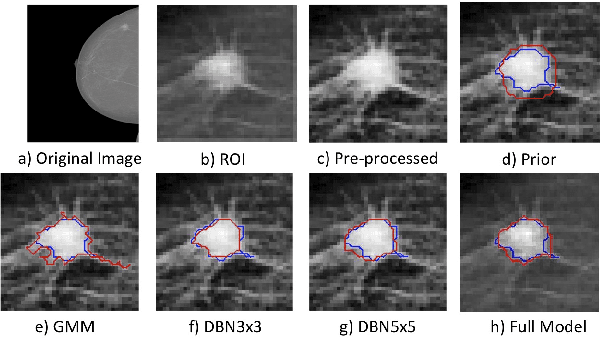
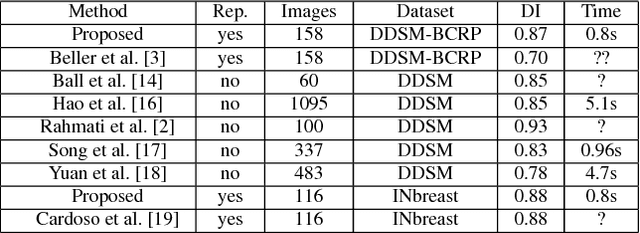
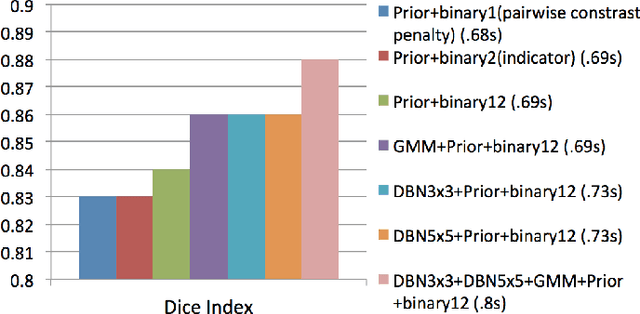
Abstract:In this paper, we present a novel method for the segmentation of breast masses from mammograms exploring structured and deep learning. Specifically, using structured support vector machine (SSVM), we formulate a model that combines different types of potential functions, including one that classifies image regions using deep learning. Our main goal with this work is to show the accuracy and efficiency improvements that these relatively new techniques can provide for the segmentation of breast masses from mammograms. We also propose an easily reproducible quantitative analysis to as- sess the performance of breast mass segmentation methodologies based on widely accepted accuracy and running time measurements on public datasets, which will facilitate further comparisons for this segmentation problem. In particular, we use two publicly available datasets (DDSM-BCRP and INbreast) and propose the computa- tion of the running time taken for the methodology to produce a mass segmentation given an input image and the use of the Dice index to quantitatively measure the segmentation accuracy. For both databases, we show that our proposed methodology produces competitive results in terms of accuracy and running time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge