Nasrin Akhter
Ovarian Cancer Prediction from Ovarian Cysts Based on TVUS Using Machine Learning Algorithms
Aug 30, 2021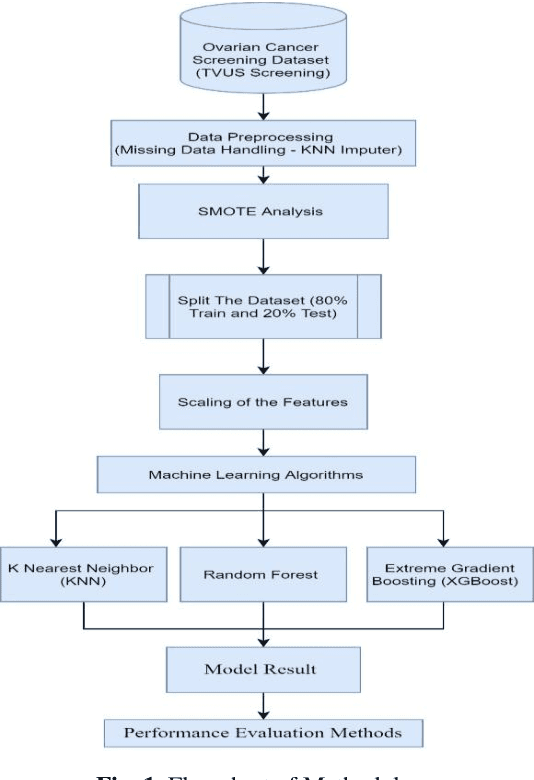
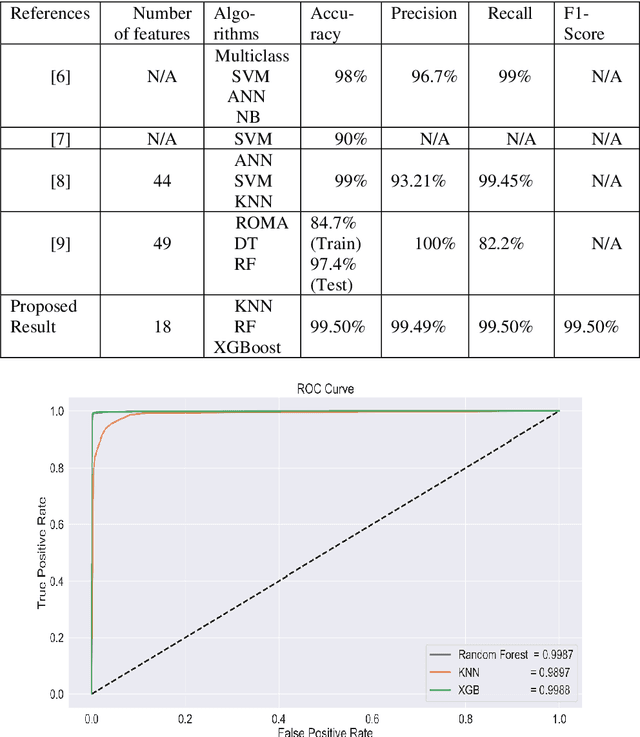
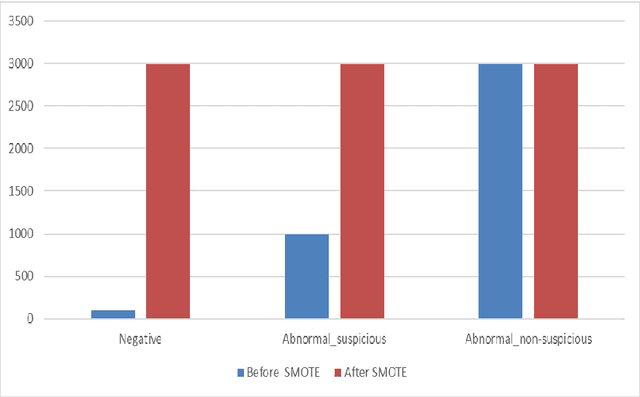
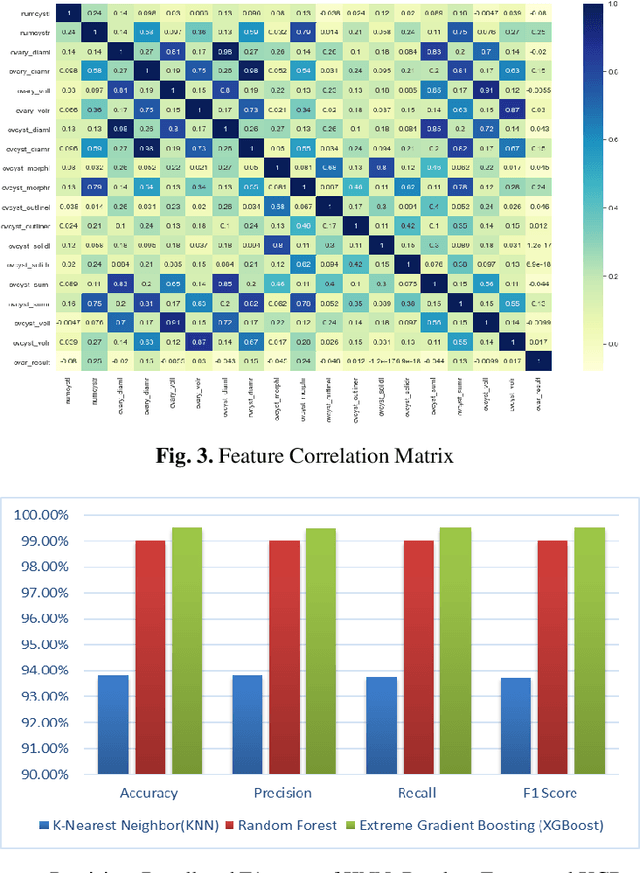
Abstract:Ovarian Cancer (OC) is type of female reproductive malignancy which can be found among young girls and mostly the women in their fertile or reproductive. There are few number of cysts are dangerous and may it cause cancer. So, it is very important to predict and it can be from different types of screening are used for this detection using Transvaginal Ultrasonography (TVUS) screening. In this research, we employed an actual datasets called PLCO with TVUS screening and three machine learning (ML) techniques, respectively Random Forest KNN, and XGBoost within three target variables. We obtained a best performance from this algorithms as far as accuracy, recall, f1 score and precision with the approximations of 99.50%, 99.50%, 99.49% and 99.50% individually. The AUC score of 99.87%, 98.97% and 99.88% are observed in these Random Forest, KNN and XGB algorithms .This approach helps assist physicians and suspects in identifying ovarian risks early on, reducing ovarian malignancy-related complications and deaths.
Decoy Selection for Protein Structure Prediction Via Extreme Gradient Boosting and Ranking
Oct 03, 2020

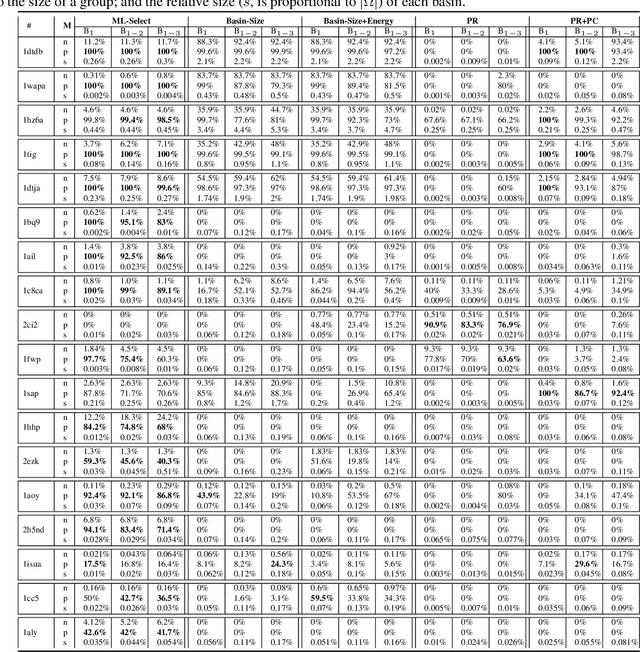
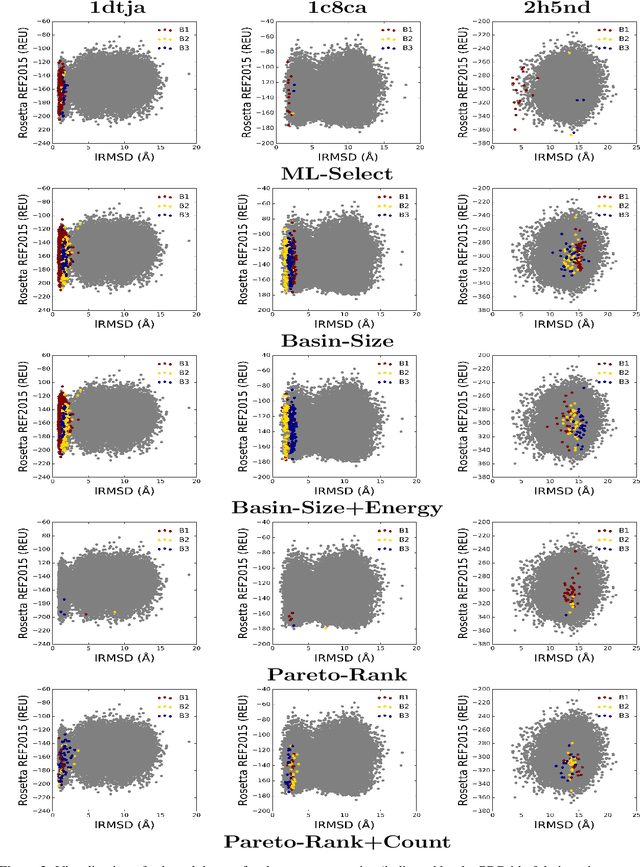
Abstract:Identifying one or more biologically-active/native decoys from millions of non-native decoys is one of the major challenges in computational structural biology. The extreme lack of balance in positive and negative samples (native and non-native decoys) in a decoy set makes the problem even more complicated. Consensus methods show varied success in handling the challenge of decoy selection despite some issues associated with clustering large decoy sets and decoy sets that do not show much structural similarity. Recent investigations into energy landscape-based decoy selection approaches show promises. However, lack of generalization over varied test cases remains a bottleneck for these methods. We propose a novel decoy selection method, ML-Select, a machine learning framework that exploits the energy landscape associated with the structure space probed through a template-free decoy generation. The proposed method outperforms both clustering and energy ranking-based methods, all the while consistently offering better performance on varied test-cases. Moreover, ML-Select shows promising results even for the decoy sets consisting of mostly low-quality decoys. ML-Select is a useful method for decoy selection. This work suggests further research in finding more effective ways to adopt machine learning frameworks in achieving robust performance for decoy selection in template-free protein structure prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge