Nadia Shakoor
Classification and Visualization of Genotype x Phenotype Interactions in Biomass Sorghum
Aug 09, 2021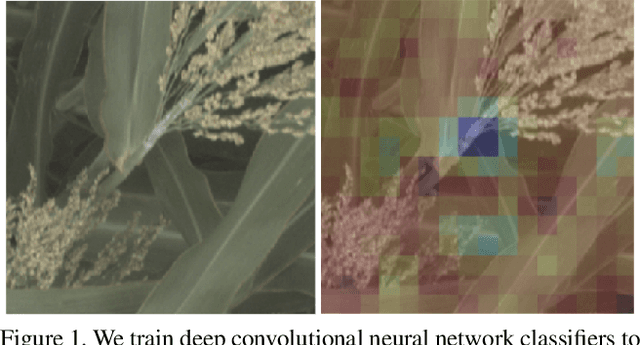



Abstract:We introduce a simple approach to understanding the relationship between single nucleotide polymorphisms (SNPs), or groups of related SNPs, and the phenotypes they control. The pipeline involves training deep convolutional neural networks (CNNs) to differentiate between images of plants with reference and alternate versions of various SNPs, and then using visualization approaches to highlight what the classification networks key on. We demonstrate the capacity of deep CNNs at performing this classification task, and show the utility of these visualizations on RGB imagery of biomass sorghum captured by the TERRA-REF gantry. We focus on several different genetic markers with known phenotypic expression, and discuss the possibilities of using this approach to uncover genotype x phenotype relationships.
Multi-resolution Outlier Pooling for Sorghum Classification
Jun 22, 2021



Abstract:Automated high throughput plant phenotyping involves leveraging sensors, such as RGB, thermal and hyperspectral cameras (among others), to make large scale and rapid measurements of the physical properties of plants for the purpose of better understanding the difference between crops and facilitating rapid plant breeding programs. One of the most basic phenotyping tasks is to determine the cultivar, or species, in a particular sensor product. This simple phenotype can be used to detect errors in planting and to learn the most differentiating features between cultivars. It is also a challenging visual recognition task, as a large number of highly related crops are grown simultaneously, leading to a classification problem with low inter-class variance. In this paper, we introduce the Sorghum-100 dataset, a large dataset of RGB imagery of sorghum captured by a state-of-the-art gantry system, a multi-resolution network architecture that learns both global and fine-grained features on the crops, and a new global pooling strategy called Dynamic Outlier Pooling which outperforms standard global pooling strategies on this task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge