Muhammad Usman Akram
A Comprehensive Review of Artificial Intelligence Applications in Major Retinal Conditions
Nov 22, 2023
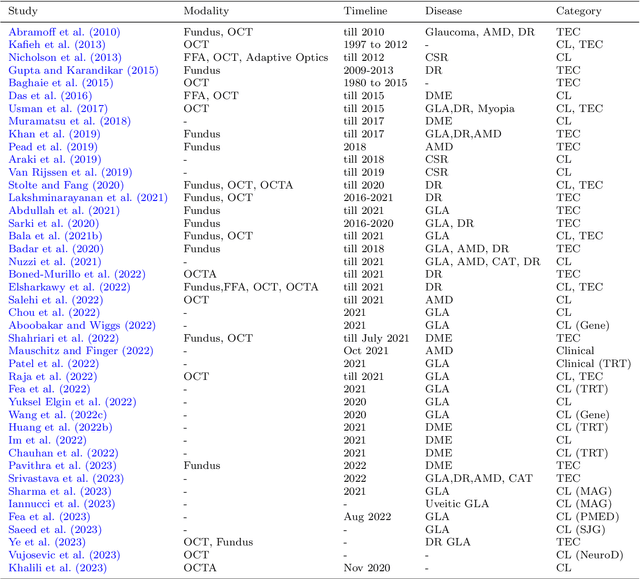
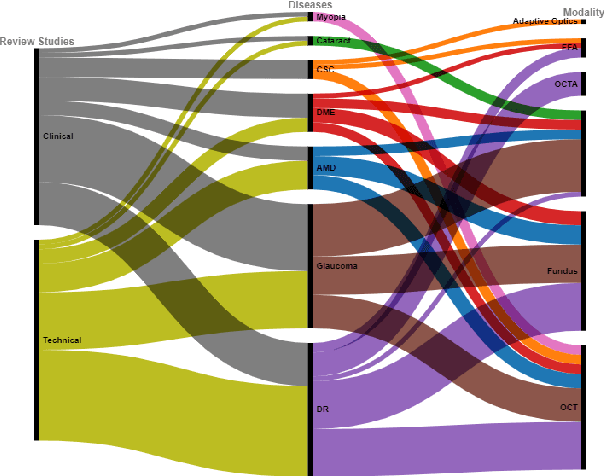
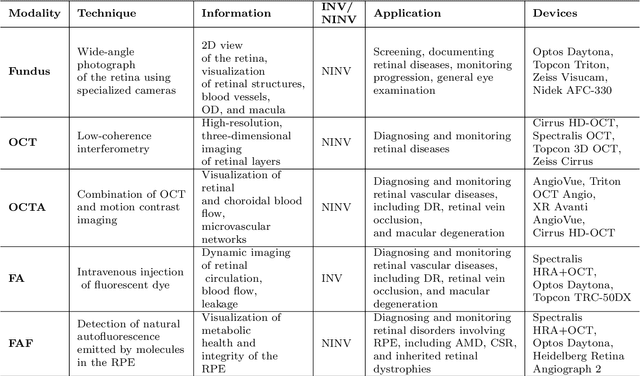
Abstract:This paper provides a systematic survey of retinal diseases that cause visual impairments or blindness, emphasizing the importance of early detection for effective treatment. It covers both clinical and automated approaches for detecting retinal disease, focusing on studies from the past decade. The survey evaluates various algorithms for identifying structural abnormalities and diagnosing retinal diseases, and it identifies future research directions based on a critical analysis of existing literature. This comprehensive study, which reviews both clinical and automated detection methods using different modalities, appears to be unique in its scope. Additionally, the survey serves as a helpful guide for researchers interested in digital retinopathy.
An Incremental Learning Approach to Automatically Recognize Pulmonary Diseases from the Multi-vendor Chest Radiographs
Jan 14, 2022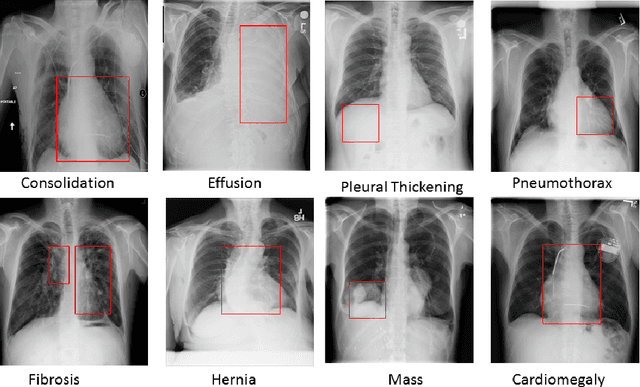
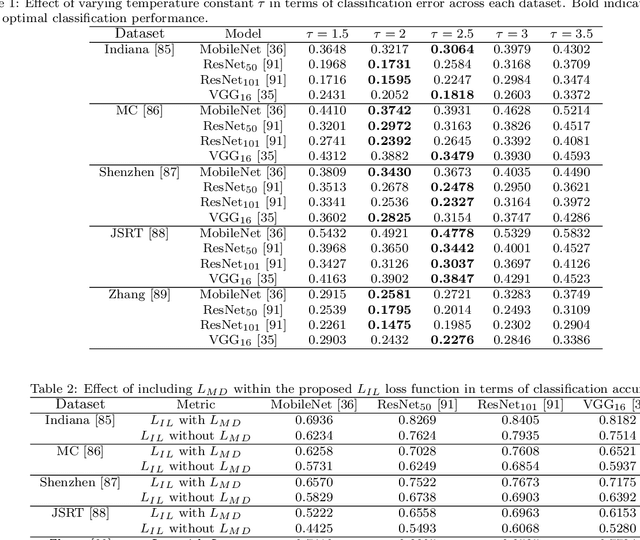
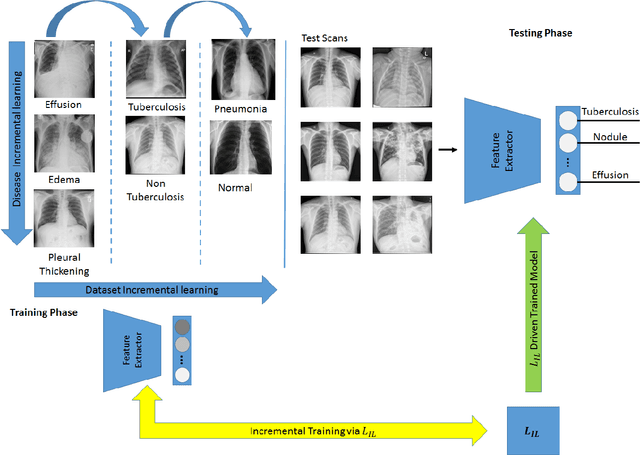
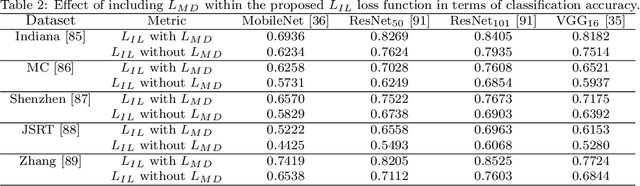
Abstract:Pulmonary diseases can cause severe respiratory problems, leading to sudden death if not treated timely. Many researchers have utilized deep learning systems to diagnose pulmonary disorders using chest X-rays (CXRs). However, such systems require exhaustive training efforts on large-scale data to effectively diagnose chest abnormalities. Furthermore, procuring such large-scale data is often infeasible and impractical, especially for rare diseases. With the recent advances in incremental learning, researchers have periodically tuned deep neural networks to learn different classification tasks with few training examples. Although, such systems can resist catastrophic forgetting, they treat the knowledge representations independently of each other, and this limits their classification performance. Also, to the best of our knowledge, there is no incremental learning-driven image diagnostic framework that is specifically designed to screen pulmonary disorders from the CXRs. To address this, we present a novel framework that can learn to screen different chest abnormalities incrementally. In addition to this, the proposed framework is penalized through an incremental learning loss function that infers Bayesian theory to recognize structural and semantic inter-dependencies between incrementally learned knowledge representations to diagnose the pulmonary diseases effectively, regardless of the scanner specifications. We tested the proposed framework on five public CXR datasets containing different chest abnormalities, where it outperformed various state-of-the-art system through various metrics.
* Computers in Biology and Medicine
Incremental Cross-Domain Adaptation for Robust Retinopathy Screening via Bayesian Deep Learning
Nov 04, 2021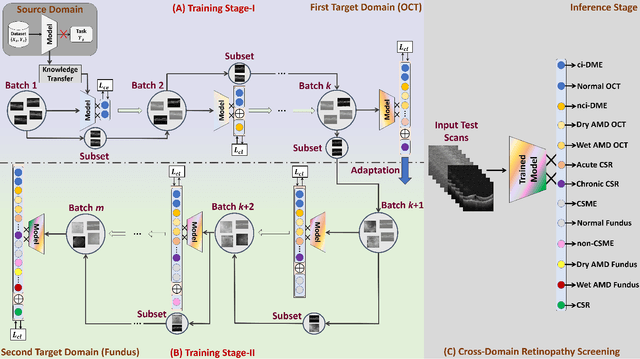
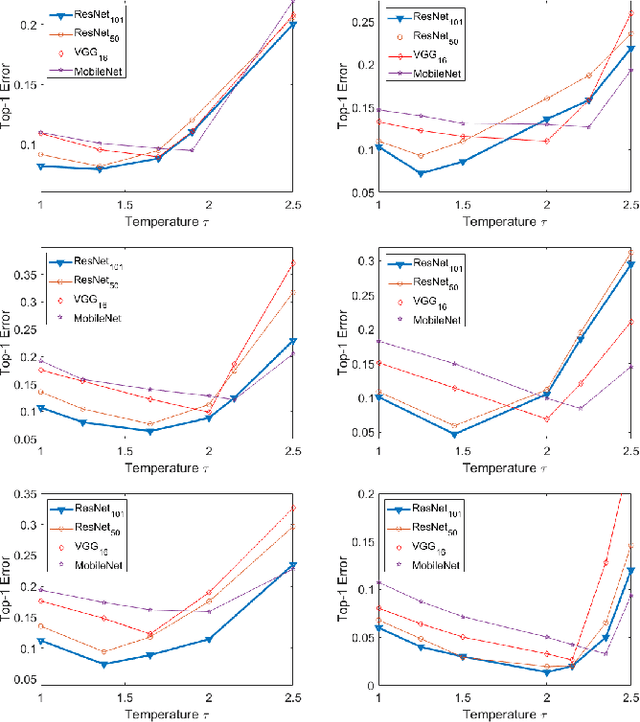
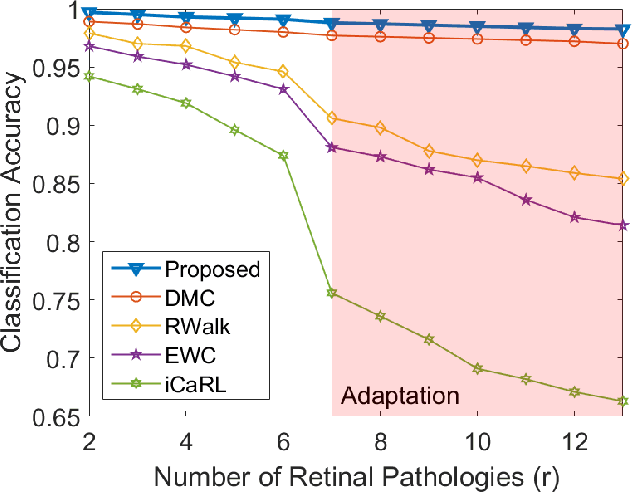
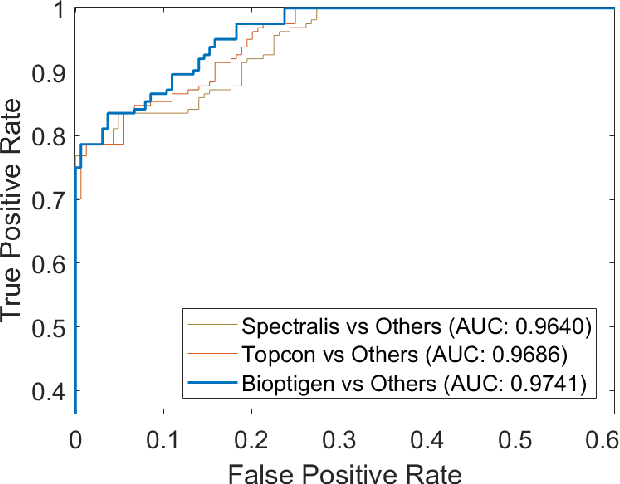
Abstract:Retinopathy represents a group of retinal diseases that, if not treated timely, can cause severe visual impairments or even blindness. Many researchers have developed autonomous systems to recognize retinopathy via fundus and optical coherence tomography (OCT) imagery. However, most of these frameworks employ conventional transfer learning and fine-tuning approaches, requiring a decent amount of well-annotated training data to produce accurate diagnostic performance. This paper presents a novel incremental cross-domain adaptation instrument that allows any deep classification model to progressively learn abnormal retinal pathologies in OCT and fundus imagery via few-shot training. Furthermore, unlike its competitors, the proposed instrument is driven via a Bayesian multi-objective function that not only enforces the candidate classification network to retain its prior learned knowledge during incremental training but also ensures that the network understands the structural and semantic relationships between previously learned pathologies and newly added disease categories to effectively recognize them at the inference stage. The proposed framework, evaluated on six public datasets acquired with three different scanners to screen thirteen retinal pathologies, outperforms the state-of-the-art competitors by achieving an overall accuracy and F1 score of 0.9826 and 0.9846, respectively.
* Accepted in IEEE Transactions on Instrumentation and Measurement. Source code is available at https://github.com/taimurhassan/continual_learning/
Clinically Verified Hybrid Deep Learning System for Retinal Ganglion Cells Aware Grading of Glaucomatous Progression
Oct 08, 2020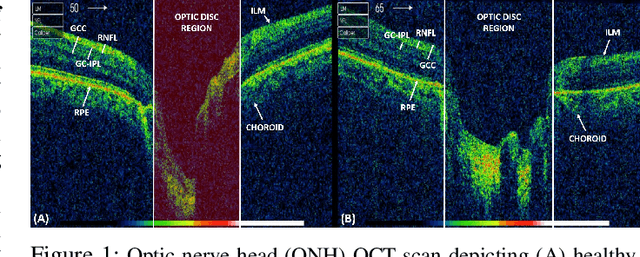
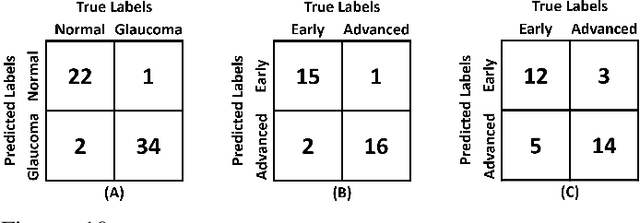
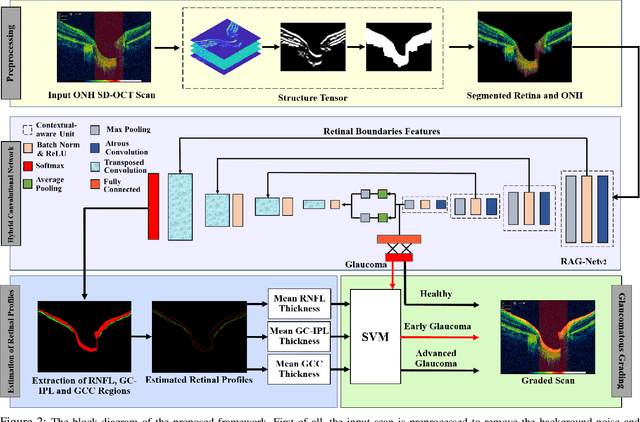
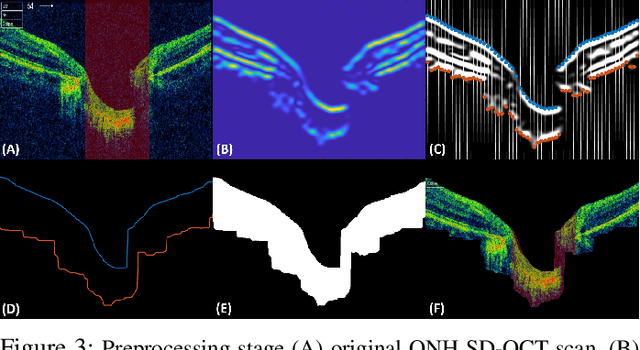
Abstract:Objective: Glaucoma is the second leading cause of blindness worldwide. Glaucomatous progression can be easily monitored by analyzing the degeneration of retinal ganglion cells (RGCs). Many researchers have screened glaucoma by measuring cup-to-disc ratios from fundus and optical coherence tomography scans. However, this paper presents a novel strategy that pays attention to the RGC atrophy for screening glaucomatous pathologies and grading their severity. Methods: The proposed framework encompasses a hybrid convolutional network that extracts the retinal nerve fiber layer, ganglion cell with the inner plexiform layer and ganglion cell complex regions, allowing thus a quantitative screening of glaucomatous subjects. Furthermore, the severity of glaucoma in screened cases is objectively graded by analyzing the thickness of these regions. Results: The proposed framework is rigorously tested on publicly available Armed Forces Institute of Ophthalmology (AFIO) dataset, where it achieved the F1 score of 0.9577 for diagnosing glaucoma, a mean dice coefficient score of 0.8697 for extracting the RGC regions and an accuracy of 0.9117 for grading glaucomatous progression. Furthermore, the performance of the proposed framework is clinically verified with the markings of four expert ophthalmologists, achieving a statistically significant Pearson correlation coefficient of 0.9236. Conclusion: An automated assessment of RGC degeneration yields better glaucomatous screening and grading as compared to the state-of-the-art solutions. Significance: An RGC-aware system not only screens glaucoma but can also grade its severity and here we present an end-to-end solution that is thoroughly evaluated on a standardized dataset and is clinically validated for analyzing glaucomatous pathologies.
Physical Action Categorization using Signal Analysis and Machine Learning
Aug 16, 2020



Abstract:Daily life of thousands of individuals around the globe suffers due to physical or mental disability related to limb movement. The quality of life for such individuals can be made better by use of assistive applications and systems. In such scenario, mapping of physical actions from movement to a computer aided application can lead the way for solution. Surface Electromyography (sEMG) presents a non-invasive mechanism through which we can translate the physical movement to signals for classification and use in applications. In this paper, we propose a machine learning based framework for classification of 4 physical actions. The framework looks into the various features from different modalities which contribution from time domain, frequency domain, higher order statistics and inter channel statistics. Next, we conducted a comparative analysis of k-NN, SVM and ELM classifier using the feature set. Effect of different combinations of feature set has also been recorded. Finally, the classifier accuracy with SVM and 1-NN based classifier for a subset of features gives an accuracy of 95.21 and 95.83 respectively. Additionally, we have also proposed that dimensionality reduction by use of PCA leads to only a minor drop of less than 5.55% in accuracy while using only 9.22% of the original feature set. These finding are useful for algorithm designer to choose the best approach keeping in mind the resources available for execution of algorithm.
Evaluation of Deep Segmentation Models for the Extraction of Retinal Lesions from Multi-modal Retinal Images
Jun 04, 2020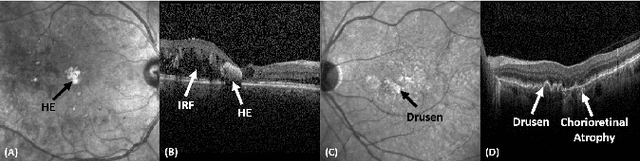
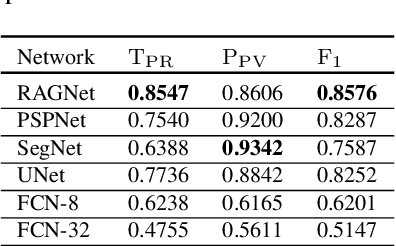
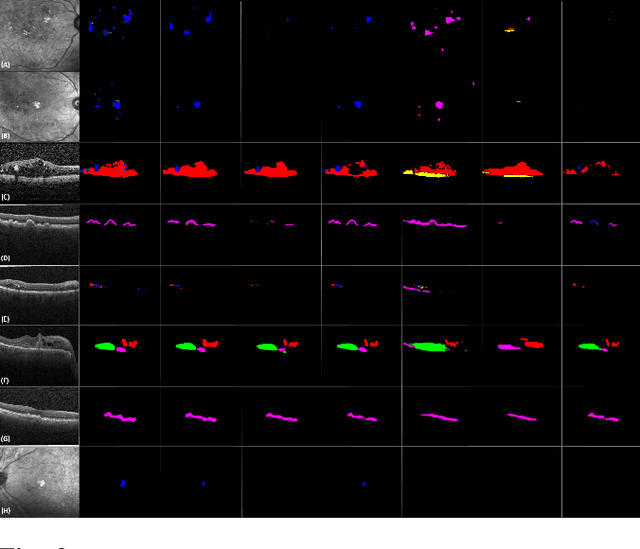
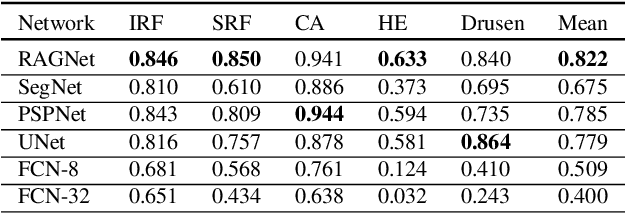
Abstract:Identification of lesions plays a vital role in the accurate classification of retinal diseases and in helping clinicians analyzing the disease severity. In this paper, we present a detailed evaluation of RAGNet, PSPNet, SegNet, UNet, FCN-8 and FCN-32 for the extraction of retinal lesions such as intra-retinal fluid, sub-retinal fluid, hard exudates, drusen, and other chorioretinal anomalies from retinal fundus and OCT scans. We also discuss the transferability of these models for extracting retinal lesions by varying training-testing dataset pairs. A total of 363 fundus and 173,915 OCT scans were considered in this evaluation from seven publicly available datasets from which 297 fundus and 59,593 OCT scans were used for testing purposes. Overall, the best performance is achieved by RAGNet with a mean dice coefficient ($\mathrm{D_C}$) score of 0.822 for extracting retinal lesions. The second-best performance is achieved by PSPNet (mean $\mathrm{D_C}$: 0.785) using ResNet\textsubscript{50} as a backbone. Moreover, the best performance for extracting drusen is achieved by UNet ($\mathrm{D_C}$: 0.864). The source code is available at: http://biomisa.org/index.php/downloads/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge