Muhammad Saif-ur-Rehman
Understanding Activation Patterns in Artificial Neural Networks by Exploring Stochastic Processes
Aug 01, 2023Abstract:To gain a deeper understanding of the behavior and learning dynamics of (deep) artificial neural networks, it is valuable to employ mathematical abstractions and models. These tools provide a simplified perspective on network performance and facilitate systematic investigations through simulations. In this paper, we propose utilizing the framework of stochastic processes, which has been underutilized thus far. Our approach models activation patterns of thresholded nodes in (deep) artificial neural networks as stochastic processes. We focus solely on activation frequency, leveraging neuroscience techniques used for real neuron spike trains. During a classification task, we extract spiking activity and use an arrival process following the Poisson distribution. We examine observed data from various artificial neural networks in image recognition tasks, fitting the proposed model's assumptions. Through this, we derive parameters describing activation patterns in each network. Our analysis covers randomly initialized, generalizing, and memorizing networks, revealing consistent differences across architectures and training sets. Calculating Mean Firing Rate, Mean Fano Factor, and Variances, we find stable indicators of memorization during learning, providing valuable insights into network behavior. The proposed model shows promise in describing activation patterns and could serve as a general framework for future investigations. It has potential applications in theoretical simulations, pruning, and transfer learning.
Adaptive SpikeDeep-Classifier: Self-organizing and self-supervised machine learning algorithm for online spike sorting
Mar 30, 2023



Abstract:Objective. Research on brain-computer interfaces (BCIs) is advancing towards rehabilitating severely disabled patients in the real world. Two key factors for successful decoding of user intentions are the size of implanted microelectrode arrays and a good online spike sorting algorithm. A small but dense microelectrode array with 3072 channels was recently developed for decoding user intentions. The process of spike sorting determines the spike activity (SA) of different sources (neurons) from recorded neural data. Unfortunately, current spike sorting algorithms are unable to handle the massively increasing amount of data from dense microelectrode arrays, making spike sorting a fragile component of the online BCI decoding framework. Approach. We proposed an adaptive and self-organized algorithm for online spike sorting, named Adaptive SpikeDeep-Classifier (Ada-SpikeDeepClassifier), which uses SpikeDeeptector for channel selection, an adaptive background activity rejector (Ada-BAR) for discarding background events, and an adaptive spike classifier (Ada-Spike classifier) for classifying the SA of different neural units. Results. Our algorithm outperformed our previously published SpikeDeep-Classifier and eight other spike sorting algorithms, as evaluated on a human dataset and a publicly available simulated dataset. Significance. The proposed algorithm is the first spike sorting algorithm that automatically learns the abrupt changes in the distribution of noise and SA. It is an artificial neural network-based algorithm that is well-suited for hardware implementation on neuromorphic chips that can be used for wearable invasive BCIs.
Invariance to Quantile Selection in Distributional Continuous Control
Dec 29, 2022Abstract:In recent years distributional reinforcement learning has produced many state of the art results. Increasingly sample efficient Distributional algorithms for the discrete action domain have been developed over time that vary primarily in the way they parameterize their approximations of value distributions, and how they quantify the differences between those distributions. In this work we transfer three of the most well-known and successful of those algorithms (QR-DQN, IQN and FQF) to the continuous action domain by extending two powerful actor-critic algorithms (TD3 and SAC) with distributional critics. We investigate whether the relative performance of the methods for the discrete action space translates to the continuous case. To that end we compare them empirically on the pybullet implementations of a set of continuous control tasks. Our results indicate qualitative invariance regarding the number and placement of distributional atoms in the deterministic, continuous action setting.
ConTraNet: A single end-to-end hybrid network for EEG-based and EMG-based human machine interfaces
Jun 21, 2022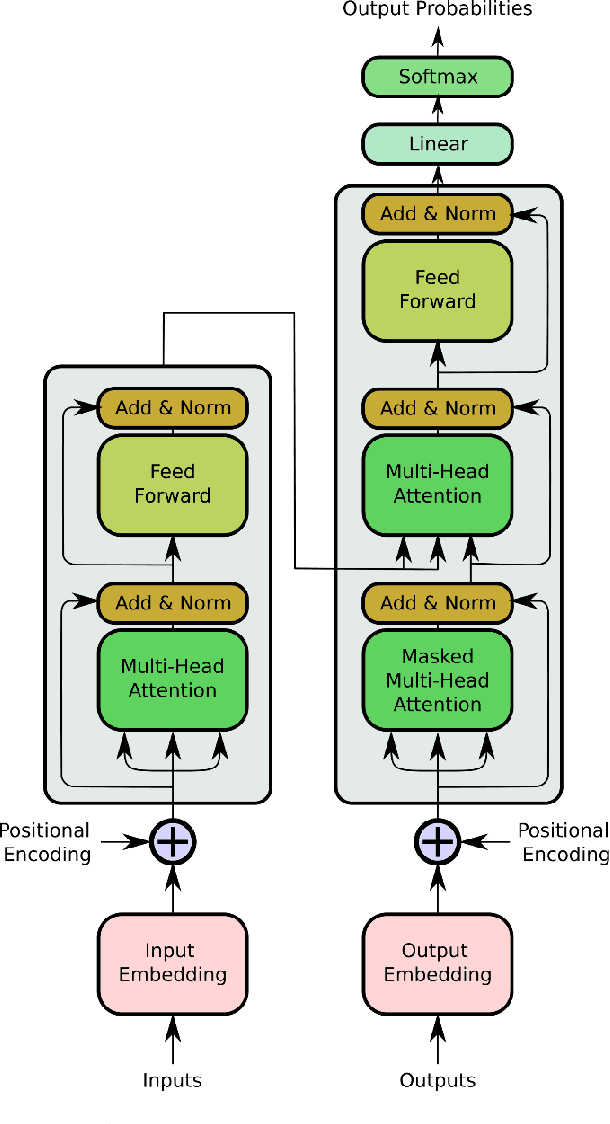
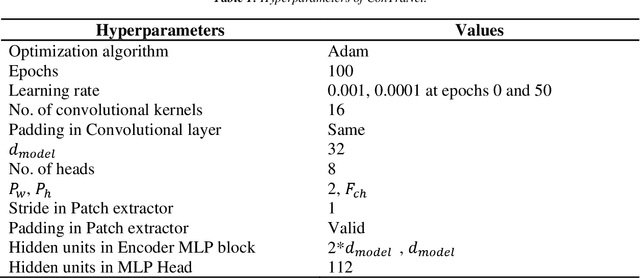
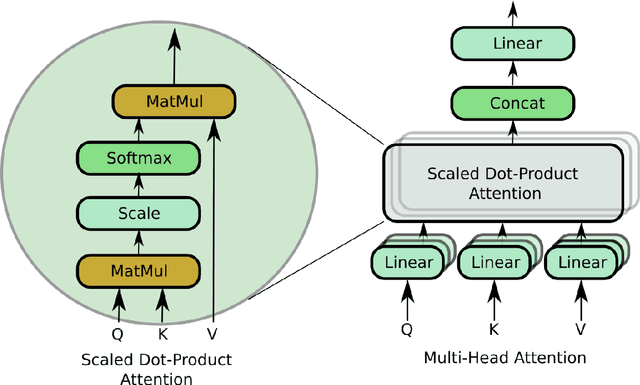
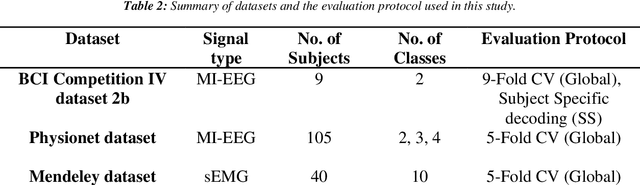
Abstract:Objective: Electroencephalography (EEG) and electromyography (EMG) are two non-invasive bio-signals, which are widely used in human machine interface (HMI) technologies (EEG-HMI and EMG-HMI paradigm) for the rehabilitation of physically disabled people. Successful decoding of EEG and EMG signals into respective control command is a pivotal step in the rehabilitation process. Recently, several Convolutional neural networks (CNNs) based architectures are proposed that directly map the raw time-series signal into decision space and the process of meaningful features extraction and classification are performed simultaneously. However, these networks are tailored to the learn the expected characteristics of the given bio-signal and are limited to single paradigm. In this work, we addressed the question that can we build a single architecture which is able to learn distinct features from different HMI paradigms and still successfully classify them. Approach: In this work, we introduce a single hybrid model called ConTraNet, which is based on CNN and Transformer architectures that is equally useful for EEG-HMI and EMG-HMI paradigms. ConTraNet uses CNN block to introduce inductive bias in the model and learn local dependencies, whereas the Transformer block uses the self-attention mechanism to learn the long-range dependencies in the signal, which are crucial for the classification of EEG and EMG signals. Main results: We evaluated and compared the ConTraNet with state-of-the-art methods on three publicly available datasets which belong to EEG-HMI and EMG-HMI paradigms. ConTraNet outperformed its counterparts in all the different category tasks (2-class, 3-class, 4-class, and 10-class decoding tasks). Significance: The results suggest that ConTraNet is robust to learn distinct features from different HMI paradigms and generalizes well as compared to the current state of the art algorithms.
Deep Transfer-Learning for patient specific model re-calibration: Application to sEMG-Classification
Dec 30, 2021
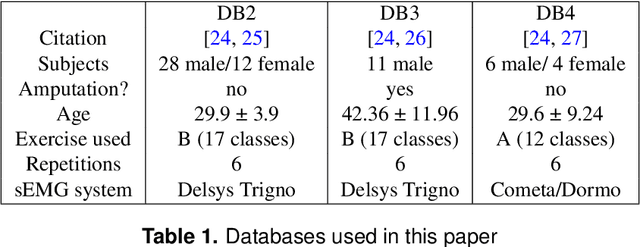

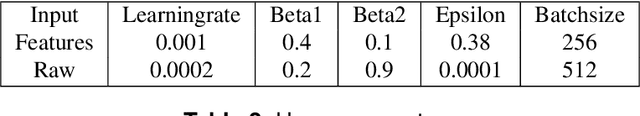
Abstract:Accurate decoding of surface electromyography (sEMG) is pivotal for muscle-to-machine-interfaces (MMI) and their application for e.g. rehabilitation therapy. sEMG signals have high inter-subject variability, due to various factors, including skin thickness, body fat percentage, and electrode placement. Therefore, obtaining high generalization quality of a trained sEMG decoder is quite challenging. Usually, machine learning based sEMG decoders are either trained on subject-specific data, or at least recalibrated for each user, individually. Even though, deep learning algorithms produced several state of the art results for sEMG decoding,however, due to the limited amount of availability of sEMG data, the deep learning models are prone to overfitting. Recently, transfer learning for domain adaptation improved generalization quality with reduced training time on various machine learning tasks. In this study, we investigate the effectiveness of transfer learning using weight initialization for recalibration of two different pretrained deep learning models on a new subjects data, and compare their performance to subject-specific models. To the best of our knowledge, this is the first study that thoroughly investigated weight-initialization based transfer learning for sEMG classification and compared transfer learning to subject-specific modeling. We tested our models on three publicly available databases under various settings. On average over all settings, our transfer learning approach improves 5~\%-points on the pretrained models without fine-tuning and 12~\%-points on the subject-specific models, while being trained on average 22~\% fewer epochs. Our results indicate that transfer learning enables faster training on fewer samples than user-specific models, and improves the performance of pretrained models as long as enough data is available.
Improving the performance of EEG decoding using anchored-STFT in conjunction with gradient norm adversarial augmentation
Nov 30, 2020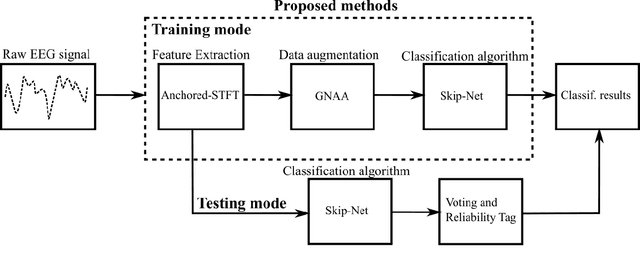
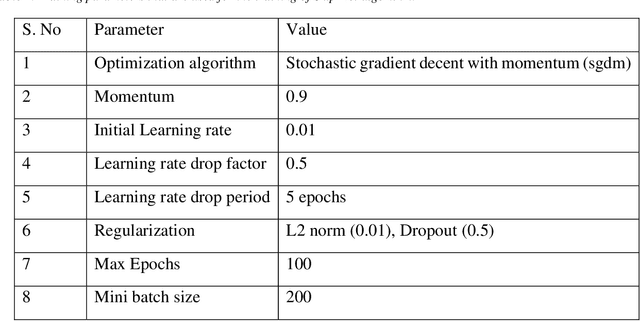

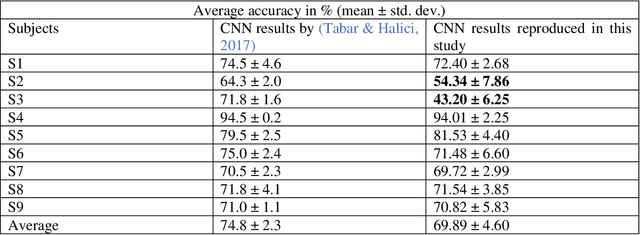
Abstract:Brain-computer interfaces (BCIs) enable direct communication between humans and machines by translating brain activity into control commands. EEG is one of the most common sources of neural signals because of its inexpensive and non-invasive nature. However, interpretation of EEG signals is non-trivial because EEG signals have a low spatial resolution and are often distorted with noise and artifacts. Therefore, it is possible that meaningful patterns for classifying EEG signals are deeply hidden. Nowadays, state-of-the-art deep-learning algorithms have proven to be quite efficient in learning hidden, meaningful patterns. However, the performance of the deep learning algorithms depends upon the quality and the amount of the provided training data. Hence, a better input formation (feature extraction) technique and a generative model to produce high-quality data can enable the deep learning algorithms to adapt high generalization quality. In this study, we proposed a novel input formation (feature extraction) method in conjunction with a novel deep learning based generative model to harness new training examples. The feature vectors are extracted using a modified Short Time Fourier Transform (STFT) called anchored-STFT. Anchored-STFT, inspired by wavelet transform, tries to minimize the tradeoff between time and frequency resolution. As a result, it extracts the inputs (feature vectors) with better time and frequency resolution compared to the standard STFT. Secondly, we introduced a novel generative adversarial data augmentation technique called gradient norm adversarial augmentation (GNAA) for generating more training data. Thirdly, we investigated the existence and significance of adversarial inputs in EEG data. Our approach obtained the kappa value of 0.814 for BCI competition II dataset III and 0.755 for BCI competition IV dataset 2b for session-to-session transfer on test data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge