Mohd Usama
Fine-Tuning Cycle-GAN for Domain Adaptation of MRI Images
Jan 18, 2026Abstract:Magnetic Resonance Imaging (MRI) scans acquired from different scanners or institutions often suffer from domain shifts owing to variations in hardware, protocols, and acquisition parameters. This discrepancy degrades the performance of deep learning models trained on source domain data when applied to target domain images. In this study, we propose a Cycle-GAN-based model for unsupervised medical-image domain adaptation. Leveraging CycleGANs, our model learns bidirectional mappings between the source and target domains without paired training data, preserving the anatomical content of the images. By leveraging Cycle-GAN capabilities with content and disparity loss for adaptation tasks, we ensured image-domain adaptation while maintaining image integrity. Several experiments on MRI datasets demonstrated the efficacy of our model in bidirectional domain adaptation without labelled data. Furthermore, research offers promising avenues for improving the diagnostic accuracy of healthcare. The statistical results confirm that our approach improves model performance and reduces domain-related variability, thus contributing to more precise and consistent medical image analysis.
Domain Adaptation of Carotid Ultrasound Images using Generative Adversarial Network
Jan 04, 2026Abstract:Deep learning has been extensively used in medical imaging applications, assuming that the test and training datasets belong to the same probability distribution. However, a common challenge arises when working with medical images generated by different systems or even the same system with different parameter settings. Such images contain diverse textures and reverberation noise that violate the aforementioned assumption. Consequently, models trained on data from one device or setting often struggle to perform effectively with data from other devices or settings. In addition, retraining models for each specific device or setting is labor-intensive and costly. To address these issues in ultrasound images, we propose a novel Generative Adversarial Network (GAN)-based model. We formulated the domain adaptation tasks as an image-to-image translation task, in which we modified the texture patterns and removed reverberation noise in the test data images from the source domain to align with those in the target domain images while keeping the image content unchanged. We applied the proposed method to two datasets containing carotid ultrasound images from three different domains. The experimental results demonstrate that the model successfully translated the texture pattern of images and removed reverberation noise from the ultrasound images. Furthermore, we evaluated the CycleGAN approaches for a comparative study with the proposed model. The experimental findings conclusively demonstrated that the proposed model achieved domain adaptation (histogram correlation (0.960 (0.019), & 0.920 (0.043) and bhattacharya distance (0.040 (0.020), & 0.085 (0.048)), compared to no adaptation (0.916 (0.062) & 0.890 (0.077), 0.090 (0.070) & 0.121 (0.095)) for both datasets.
A Domain Adaptation Model for Carotid Ultrasound: Image Harmonization, Noise Reduction, and Impact on Cardiovascular Risk Markers
Jul 06, 2024Abstract:Deep learning has been used extensively for medical image analysis applications, assuming the training and test data adhere to the same probability distributions. However, a common challenge arises when dealing with medical images generated by different systems or even the same system with varying parameter settings. Such images often contain diverse textures and noise patterns, violating the assumption. Consequently, models trained on data from one machine or setting usually struggle to perform effectively on data from another. To address this issue in ultrasound images, we proposed a Generative Adversarial Network (GAN) based model in this paper. We formulated image harmonization and denoising tasks as an image-to-image translation task, wherein we modified the texture pattern and reduced noise in Carotid ultrasound images while keeping the image content (the anatomy) unchanged. The performance was evaluated using feature distribution and pixel-space similarity metrics. In addition, blood-to-tissue contrast and influence on computed risk markers (Gray scale median, GSM) were evaluated. The results showed that domain adaptation was achieved in both tasks (histogram correlation 0.920 and 0.844), as compared to no adaptation (0.890 and 0.707), and that the anatomy of the images was retained (structure similarity index measure of the arterial wall 0.71 and 0.80). In addition, the image noise level (contrast) did not change in the image harmonization task (-34.1 vs 35.2 dB) but was improved in the noise reduction task (-23.5 vs -46.7 dB). The model outperformed the CycleGAN in both tasks. Finally, the risk marker GSM increased by 7.6 (p<0.001) in task 1 but not in task 2. We conclude that domain translation models are powerful tools for ultrasound image improvement while retaining the underlying anatomy but that downstream calculations of risk markers may be affected.
DACB-Net: Dual Attention Guided Compact Bilinear Convolution Neural Network for Skin Disease Classification
Jul 03, 2024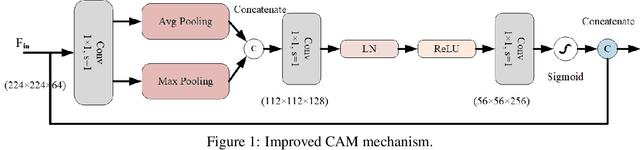
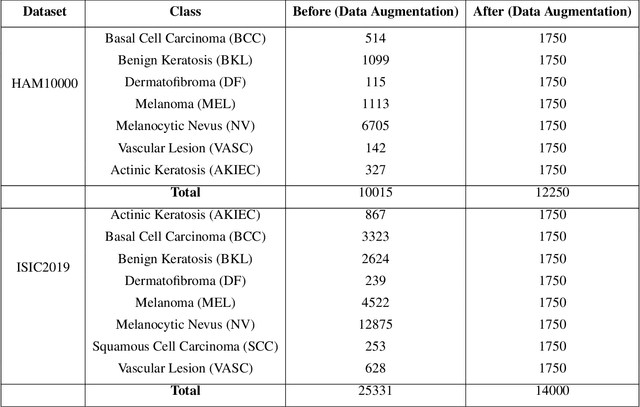
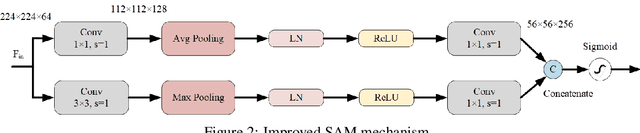
Abstract:This paper introduces the three-branch Dual Attention-Guided Compact Bilinear CNN (DACB-Net) by focusing on learning from disease-specific regions to enhance accuracy and alignment. A global branch compensates for lost discriminative features, generating Attention Heat Maps (AHM) for relevant cropped regions. Finally, the last pooling layers of global and local branches are concatenated for fine-tuning, which offers a comprehensive solution to the challenges posed by skin disease diagnosis. Although current CNNs employ Stochastic Gradient Descent (SGD) for discriminative feature learning, using distinct pairs of local image patches to compute gradients and incorporating a modulation factor in the loss for focusing on complex data during training. However, this approach can lead to dataset imbalance, weight adjustments, and vulnerability to overfitting. The proposed solution combines two supervision branches and a novel loss function to address these issues, enhancing performance and interpretability. The framework integrates data augmentation, transfer learning, and fine-tuning to tackle data imbalance to improve classification performance, and reduce computational costs. Simulations on the HAM10000 and ISIC2019 datasets demonstrate the effectiveness of this approach, showcasing a 2.59% increase in accuracy compared to the state-of-the-art.
Bilinear-Convolutional Neural Network Using a Matrix Similarity-based Joint Loss Function for Skin Disease Classification
Jun 02, 2024


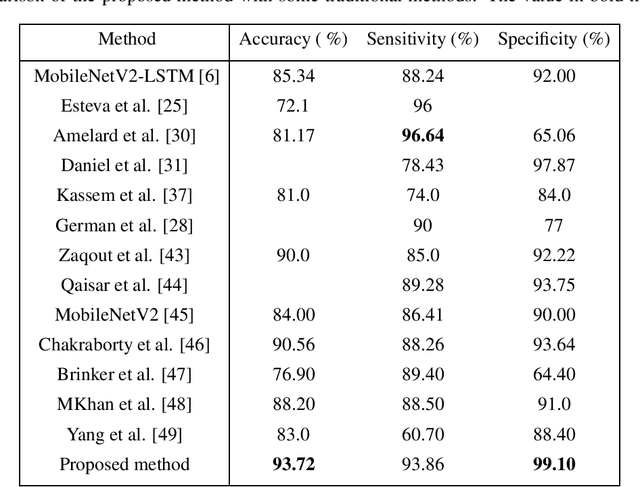
Abstract:In this study, we proposed a model for skin disease classification using a Bilinear Convolutional Neural Network (BCNN) with a Constrained Triplet Network (CTN). BCNN can capture rich spatial interactions between features in image data. This computes the outer product of feature vectors from two different CNNs by a bilinear pooling. The resulting features encode second-order statistics, enabling the network to capture more complex relationships between different channels and spatial locations. The CTN employs the Triplet Loss Function (TLF) by using a new loss layer that is added at the end of the architecture called the Constrained Triplet Loss (CTL) layer. This is done to obtain two significant learning objectives: inter-class categorization and intra-class concentration with their deep features as often as possible, which can be effective for skin disease classification. The proposed model is trained to extract the intra-class features from a deep network and accordingly increases the distance between these features, improving the model's performance. The model achieved a mean accuracy of 93.72%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge