Mohammed Safwan
BACH: Grand Challenge on Breast Cancer Histology Images
Aug 13, 2018
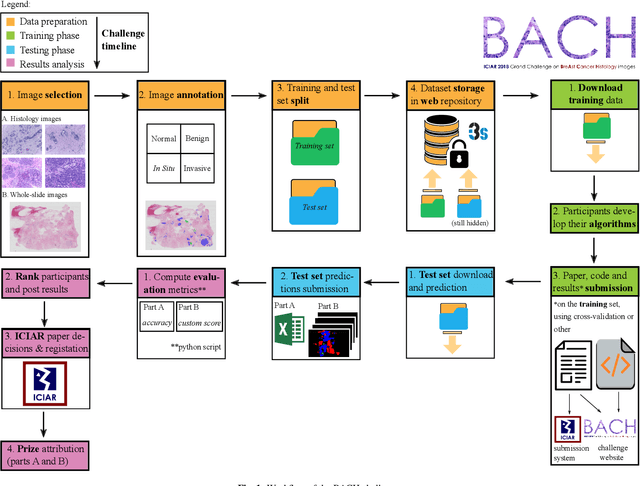
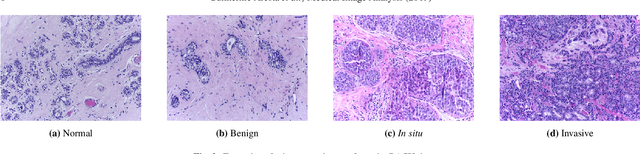
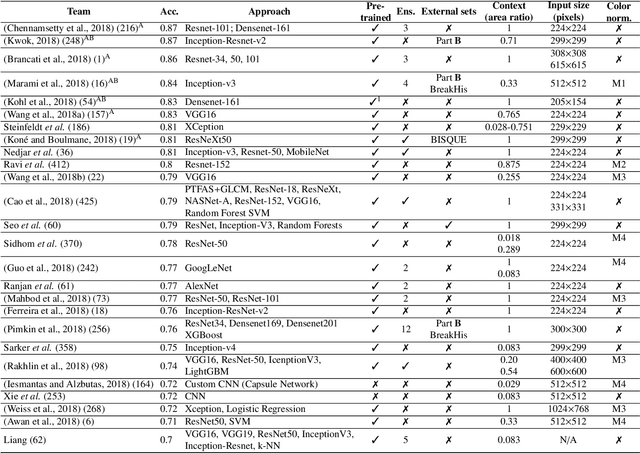
Abstract:Breast cancer is the most common invasive cancer in women, affecting more than 10% of women worldwide. Microscopic analysis of a biopsy remains one of the most important methods to diagnose the type of breast cancer. This requires specialized analysis by pathologists, in a task that i) is highly time- and cost-consuming and ii) often leads to nonconsensual results. The relevance and potential of automatic classification algorithms using hematoxylin-eosin stained histopathological images has already been demonstrated, but the reported results are still sub-optimal for clinical use. With the goal of advancing the state-of-the-art in automatic classification, the Grand Challenge on BreAst Cancer Histology images (BACH) was organized in conjunction with the 15th International Conference on Image Analysis and Recognition (ICIAR 2018). A large annotated dataset, composed of both microscopy and whole-slide images, was specifically compiled and made publicly available for the BACH challenge. Following a positive response from the scientific community, a total of 64 submissions, out of 677 registrations, effectively entered the competition. From the submitted algorithms it was possible to push forward the state-of-the-art in terms of accuracy (87%) in automatic classification of breast cancer with histopathological images. Convolutional neuronal networks were the most successful methodology in the BACH challenge. Detailed analysis of the collective results allowed the identification of remaining challenges in the field and recommendations for future developments. The BACH dataset remains publically available as to promote further improvements to the field of automatic classification in digital pathology.
Automatic Segmentation and Overall Survival Prediction in Gliomas using Fully Convolutional Neural Network and Texture Analysis
Dec 06, 2017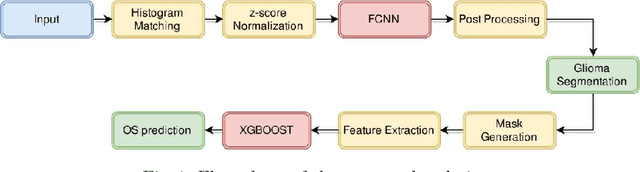
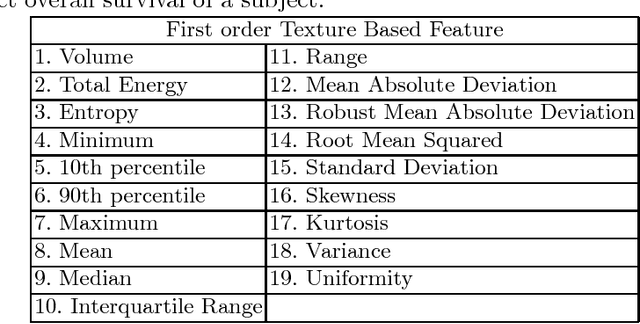
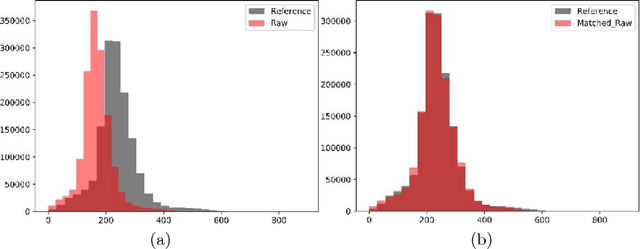
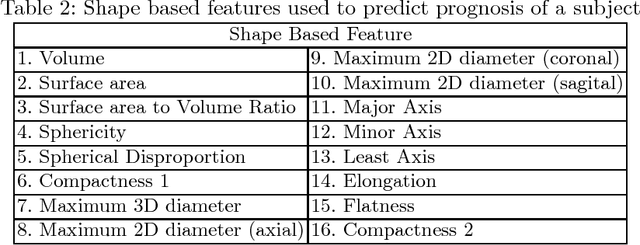
Abstract:In this paper, we use a fully convolutional neural network (FCNN) for the segmentation of gliomas from Magnetic Resonance Images (MRI). A fully automatic, voxel based classification was achieved by training a 23 layer deep FCNN on 2-D slices extracted from patient volumes. The network was trained on slices extracted from 130 patients and validated on 50 patients. For the task of survival prediction, texture and shape based features were extracted from T1 post contrast volume to train an XGBoost regressor. On BraTS 2017 validation set, the proposed scheme achieved a mean whole tumor, tumor core and active dice score of 0.83, 0.69 and 0.69 respectively and an accuracy of 52% for the overall survival prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge