Mohammad Rakibul Hasan Mahin
Med-IC: Fusing a Single Layer Involution with Convolutions for Enhanced Medical Image Classification and Segmentation
Sep 27, 2024Abstract:The majority of medical images, especially those that resemble cells, have similar characteristics. These images, which occur in a variety of shapes, often show abnormalities in the organ or cell region. The convolution operation possesses a restricted capability to extract visual patterns across several spatial regions of an image. The involution process, which is the inverse operation of convolution, complements this inherent lack of spatial information extraction present in convolutions. In this study, we investigate how applying a single layer of involution prior to a convolutional neural network (CNN) architecture can significantly improve classification and segmentation performance, with a comparatively negligible amount of weight parameters. The study additionally shows how excessive use of involution layers might result in inaccurate predictions in a particular type of medical image. According to our findings from experiments, the strategy of adding only a single involution layer before a CNN-based model outperforms most of the previous works.
Spatially Optimized Compact Deep Metric Learning Model for Similarity Search
Apr 09, 2024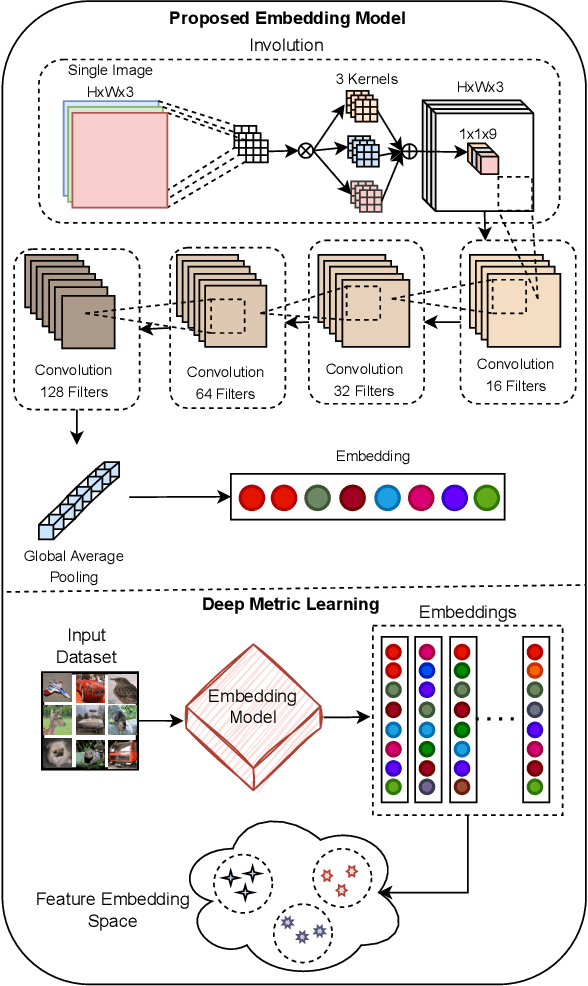
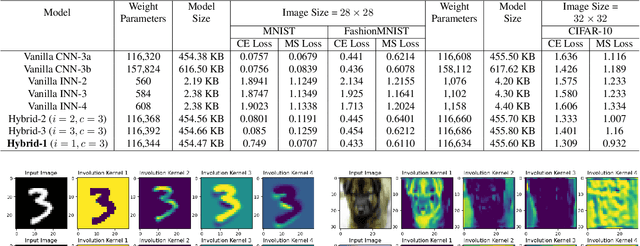
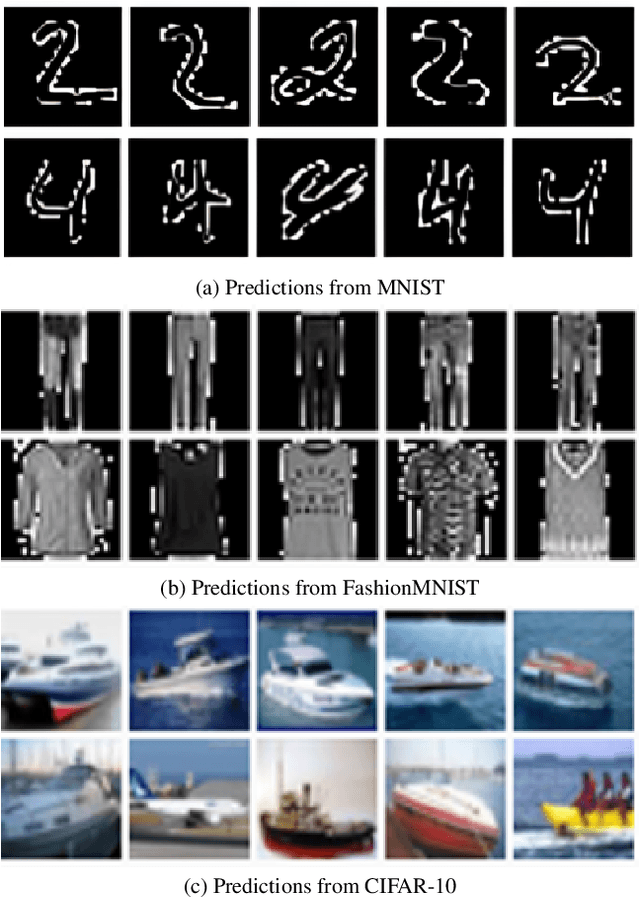
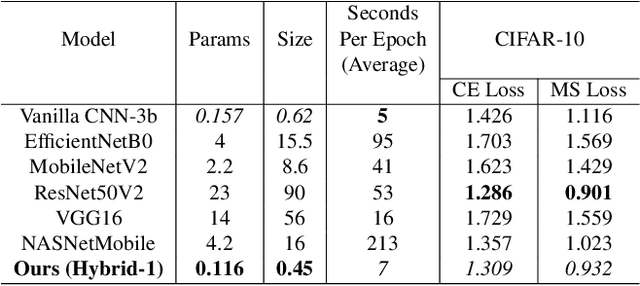
Abstract:Spatial optimization is often overlooked in many computer vision tasks. Filters should be able to recognize the features of an object regardless of where it is in the image. Similarity search is a crucial task where spatial features decide an important output. The capacity of convolution to capture visual patterns across various locations is limited. In contrast to convolution, the involution kernel is dynamically created at each pixel based on the pixel value and parameters that have been learned. This study demonstrates that utilizing a single layer of involution feature extractor alongside a compact convolution model significantly enhances the performance of similarity search. Additionally, we improve predictions by using the GELU activation function rather than the ReLU. The negligible amount of weight parameters in involution with a compact model with better performance makes the model very useful in real-world implementations. Our proposed model is below 1 megabyte in size. We have experimented with our proposed methodology and other models on CIFAR-10, FashionMNIST, and MNIST datasets. Our proposed method outperforms across all three datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge