Misaki Ohashi
FineBio: A Fine-Grained Video Dataset of Biological Experiments with Hierarchical Annotation
Feb 01, 2024



Abstract:In the development of science, accurate and reproducible documentation of the experimental process is crucial. Automatic recognition of the actions in experiments from videos would help experimenters by complementing the recording of experiments. Towards this goal, we propose FineBio, a new fine-grained video dataset of people performing biological experiments. The dataset consists of multi-view videos of 32 participants performing mock biological experiments with a total duration of 14.5 hours. One experiment forms a hierarchical structure, where a protocol consists of several steps, each further decomposed into a set of atomic operations. The uniqueness of biological experiments is that while they require strict adherence to steps described in each protocol, there is freedom in the order of atomic operations. We provide hierarchical annotation on protocols, steps, atomic operations, object locations, and their manipulation states, providing new challenges for structured activity understanding and hand-object interaction recognition. To find out challenges on activity understanding in biological experiments, we introduce baseline models and results on four different tasks, including (i) step segmentation, (ii) atomic operation detection (iii) object detection, and (iv) manipulated/affected object detection. Dataset and code are available from https://github.com/aistairc/FineBio.
Unbiased Scene Graph Generation using Predicate Similarities
Oct 03, 2022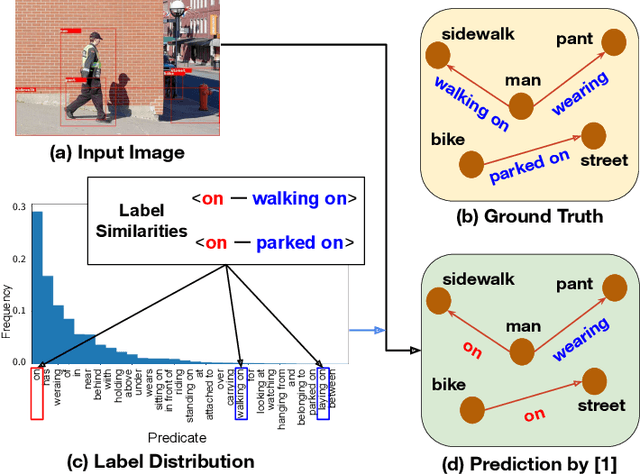



Abstract:Scene Graphs are widely applied in computer vision as a graphical representation of relationships between objects shown in images. However, these applications have not yet reached a practical stage of development owing to biased training caused by long-tailed predicate distributions. In recent years, many studies have tackled this problem. In contrast, relatively few works have considered predicate similarities as a unique dataset feature which also leads to the biased prediction. Due to the feature, infrequent predicates (e.g., parked on, covered in) are easily misclassified as closely-related frequent predicates (e.g., on, in). Utilizing predicate similarities, we propose a new classification scheme that branches the process to several fine-grained classifiers for similar predicate groups. The classifiers aim to capture the differences among similar predicates in detail. We also introduce the idea of transfer learning to enhance the features for the predicates which lack sufficient training samples to learn the descriptive representations. The results of extensive experiments on the Visual Genome dataset show that the combination of our method and an existing debiasing approach greatly improves performance on tail predicates in challenging SGCls/SGDet tasks. Nonetheless, the overall performance of the proposed approach does not reach that of the current state of the art, so further analysis remains necessary as future work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge