MingXuan Xiao
Convolutional neural network classification of cancer cytopathology images: taking breast cancer as an example
Apr 12, 2024



Abstract:Breast cancer is a relatively common cancer among gynecological cancers. Its diagnosis often relies on the pathology of cells in the lesion. The pathological diagnosis of breast cancer not only requires professionals and time, but also sometimes involves subjective judgment. To address the challenges of dependence on pathologists expertise and the time-consuming nature of achieving accurate breast pathological image classification, this paper introduces an approach utilizing convolutional neural networks (CNNs) for the rapid categorization of pathological images, aiming to enhance the efficiency of breast pathological image detection. And the approach enables the rapid and automatic classification of pathological images into benign and malignant groups. The methodology involves utilizing a convolutional neural network (CNN) model leveraging the Inceptionv3 architecture and transfer learning algorithm for extracting features from pathological images. Utilizing a neural network with fully connected layers and employing the SoftMax function for image classification. Additionally, the concept of image partitioning is introduced to handle high-resolution images. To achieve the ultimate classification outcome, the classification probabilities of each image block are aggregated using three algorithms: summation, product, and maximum. Experimental validation was conducted on the BreaKHis public dataset, resulting in accuracy rates surpassing 0.92 across all four magnification coefficients (40X, 100X, 200X, and 400X). It demonstrates that the proposed method effectively enhances the accuracy in classifying pathological images of breast cancer.
Survival Prediction Across Diverse Cancer Types Using Neural Networks
Apr 11, 2024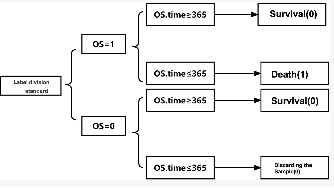
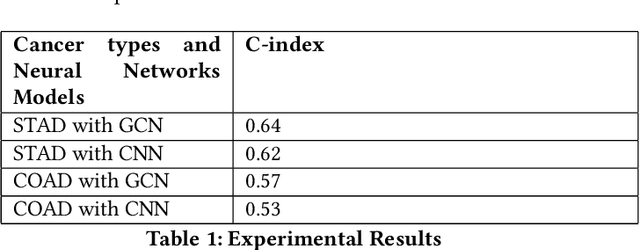

Abstract:Gastric cancer and Colon adenocarcinoma represent widespread and challenging malignancies with high mortality rates and complex treatment landscapes. In response to the critical need for accurate prognosis in cancer patients, the medical community has embraced the 5-year survival rate as a vital metric for estimating patient outcomes. This study introduces a pioneering approach to enhance survival prediction models for gastric and Colon adenocarcinoma patients. Leveraging advanced image analysis techniques, we sliced whole slide images (WSI) of these cancers, extracting comprehensive features to capture nuanced tumor characteristics. Subsequently, we constructed patient-level graphs, encapsulating intricate spatial relationships within tumor tissues. These graphs served as inputs for a sophisticated 4-layer graph convolutional neural network (GCN), designed to exploit the inherent connectivity of the data for comprehensive analysis and prediction. By integrating patients' total survival time and survival status, we computed C-index values for gastric cancer and Colon adenocarcinoma, yielding 0.57 and 0.64, respectively. Significantly surpassing previous convolutional neural network models, these results underscore the efficacy of our approach in accurately predicting patient survival outcomes. This research holds profound implications for both the medical and AI communities, offering insights into cancer biology and progression while advancing personalized treatment strategies. Ultimately, our study represents a significant stride in leveraging AI-driven methodologies to revolutionize cancer prognosis and improve patient outcomes on a global scale.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge