Michael Snyder
Stanford University
CliniDigest: A Case Study in Large Language Model Based Large-Scale Summarization of Clinical Trial Descriptions
Jul 31, 2023


Abstract:A clinical trial is a study that evaluates new biomedical interventions. To design new trials, researchers draw inspiration from those current and completed. In 2022, there were on average more than 100 clinical trials submitted to ClinicalTrials.gov every day, with each trial having a mean of approximately 1500 words [1]. This makes it nearly impossible to keep up to date. To mitigate this issue, we have created a batch clinical trial summarizer called CliniDigest using GPT-3.5. CliniDigest is, to our knowledge, the first tool able to provide real-time, truthful, and comprehensive summaries of clinical trials. CliniDigest can reduce up to 85 clinical trial descriptions (approximately 10,500 words) into a concise 200-word summary with references and limited hallucinations. We have tested CliniDigest on its ability to summarize 457 trials divided across 27 medical subdomains. For each field, CliniDigest generates summaries of $\mu=153,\ \sigma=69 $ words, each of which utilizes $\mu=54\%,\ \sigma=30\% $ of the sources. A more comprehensive evaluation is planned and outlined in this paper.
Beyond Low Earth Orbit: Biomonitoring, Artificial Intelligence, and Precision Space Health
Dec 22, 2021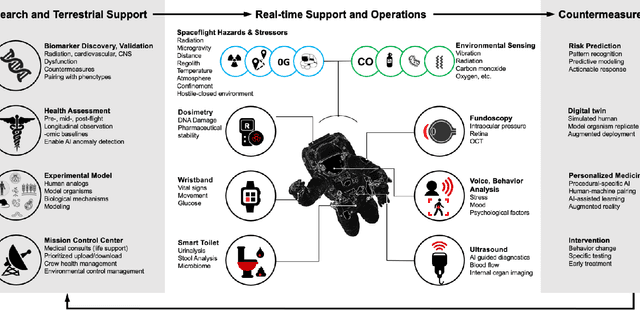
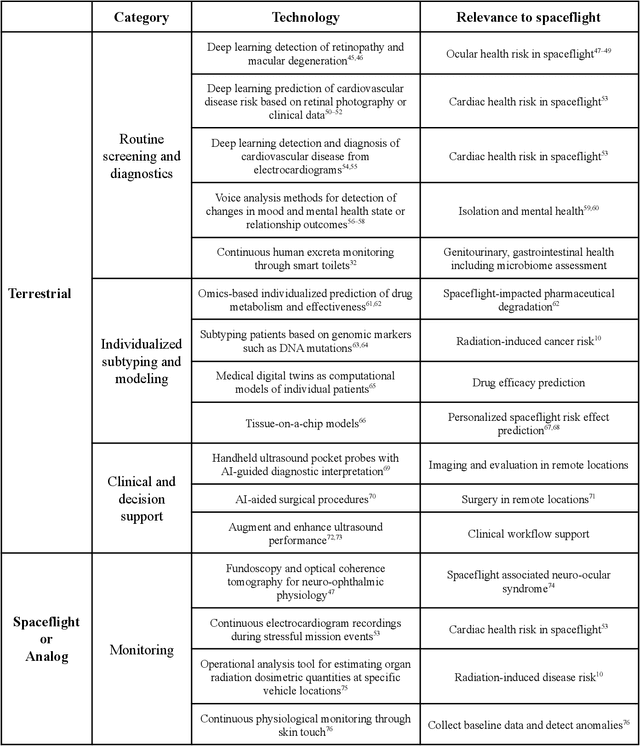
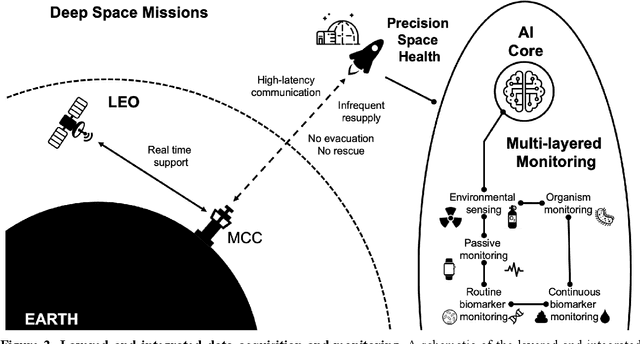
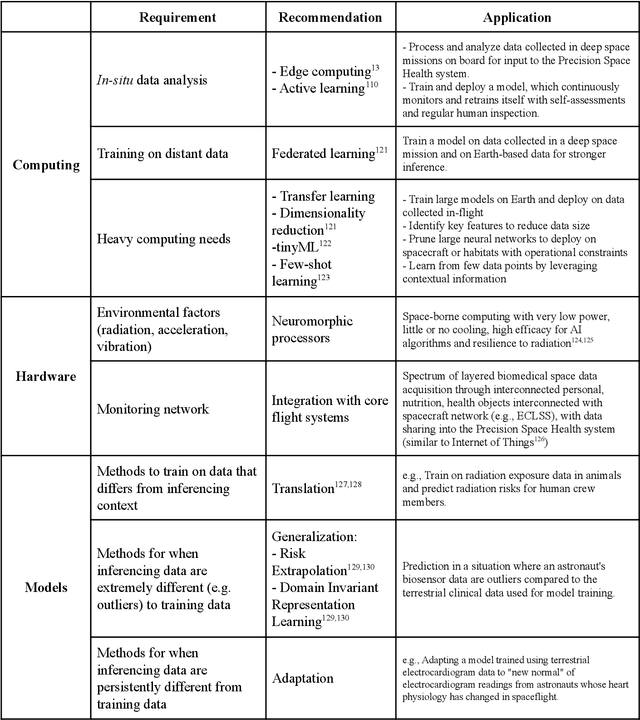
Abstract:Human space exploration beyond low Earth orbit will involve missions of significant distance and duration. To effectively mitigate myriad space health hazards, paradigm shifts in data and space health systems are necessary to enable Earth-independence, rather than Earth-reliance. Promising developments in the fields of artificial intelligence and machine learning for biology and health can address these needs. We propose an appropriately autonomous and intelligent Precision Space Health system that will monitor, aggregate, and assess biomedical statuses; analyze and predict personalized adverse health outcomes; adapt and respond to newly accumulated data; and provide preventive, actionable, and timely insights to individual deep space crew members and iterative decision support to their crew medical officer. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration, on future applications of artificial intelligence in space biology and health. In the next decade, biomonitoring technology, biomarker science, spacecraft hardware, intelligent software, and streamlined data management must mature and be woven together into a Precision Space Health system to enable humanity to thrive in deep space.
Beyond Low Earth Orbit: Biological Research, Artificial Intelligence, and Self-Driving Labs
Dec 22, 2021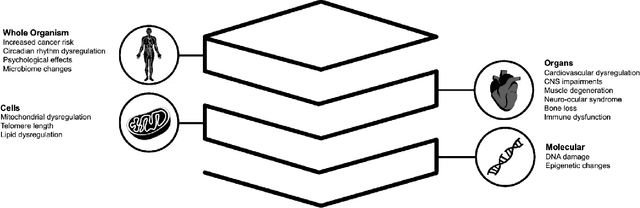
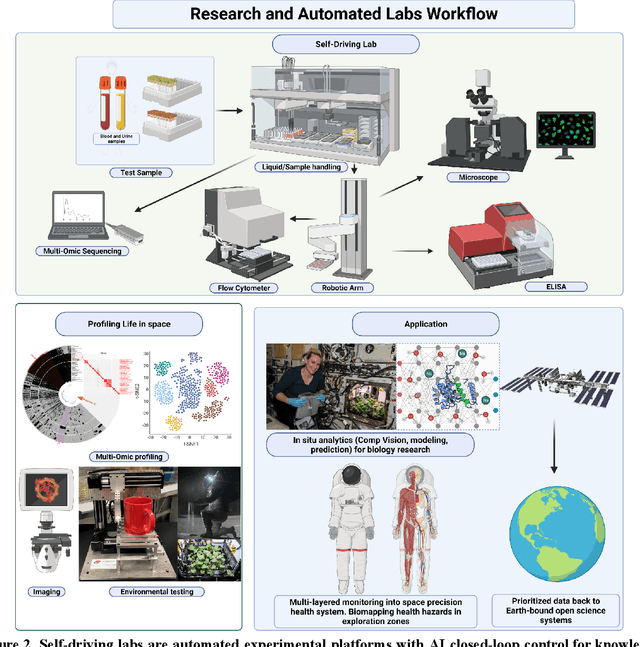
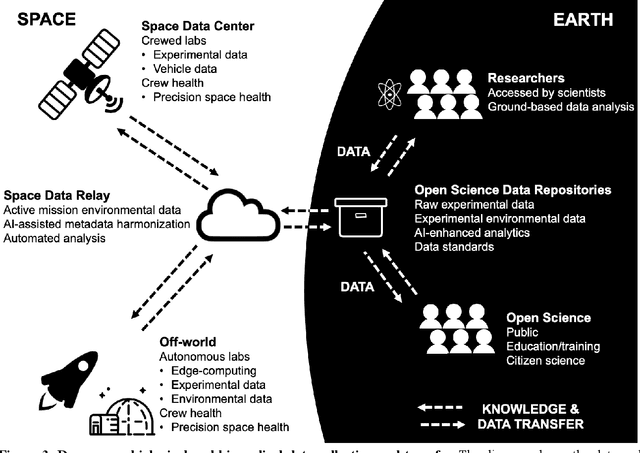
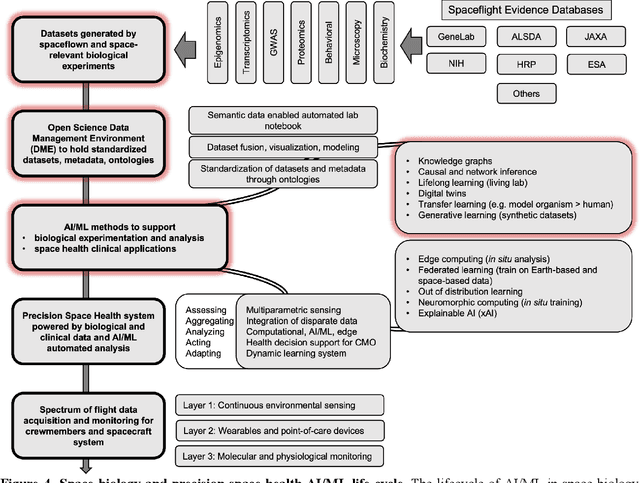
Abstract:Space biology research aims to understand fundamental effects of spaceflight on organisms, develop foundational knowledge to support deep space exploration, and ultimately bioengineer spacecraft and habitats to stabilize the ecosystem of plants, crops, microbes, animals, and humans for sustained multi-planetary life. To advance these aims, the field leverages experiments, platforms, data, and model organisms from both spaceborne and ground-analog studies. As research is extended beyond low Earth orbit, experiments and platforms must be maximally autonomous, light, agile, and intelligent to expedite knowledge discovery. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration on artificial intelligence, machine learning, and modeling applications which offer key solutions toward these space biology challenges. In the next decade, the synthesis of artificial intelligence into the field of space biology will deepen the biological understanding of spaceflight effects, facilitate predictive modeling and analytics, support maximally autonomous and reproducible experiments, and efficiently manage spaceborne data and metadata, all with the goal to enable life to thrive in deep space.
Learning Personal Food Preferences via Food Logs Embedding
Nov 22, 2021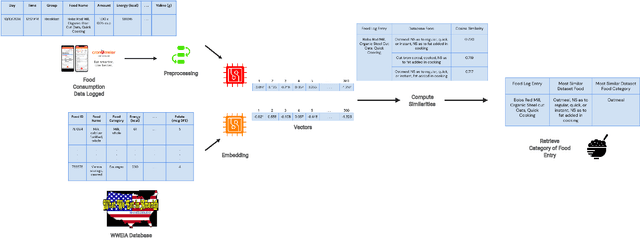
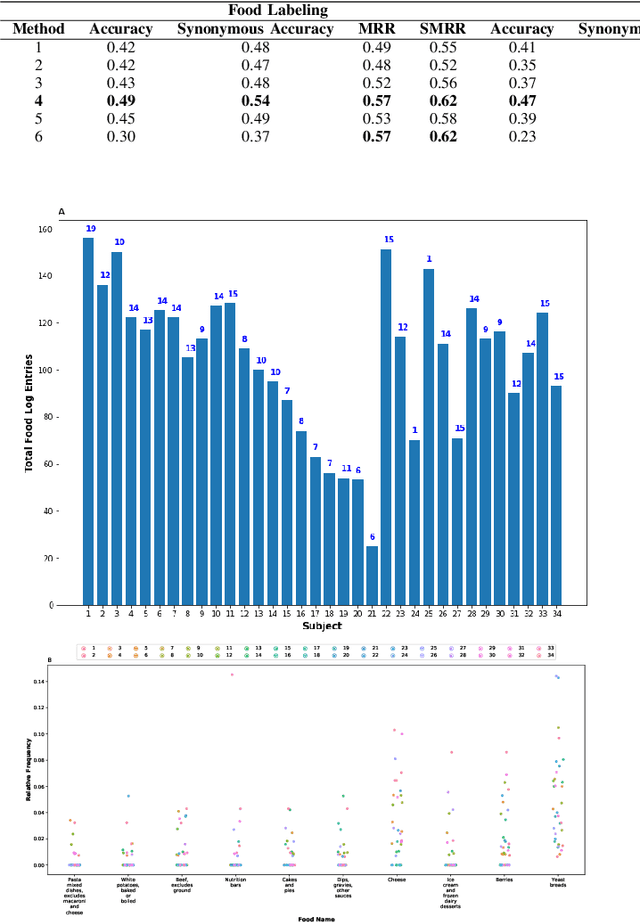
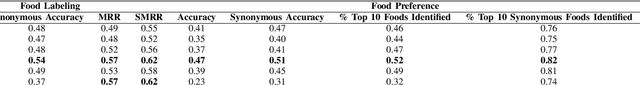
Abstract:Diet management is key to managing chronic diseases such as diabetes. Automated food recommender systems may be able to assist by providing meal recommendations that conform to a user's nutrition goals and food preferences. Current recommendation systems suffer from a lack of accuracy that is in part due to a lack of knowledge of food preferences, namely foods users like to and are able to eat frequently. In this work, we propose a method for learning food preferences from food logs, a comprehensive but noisy source of information about users' dietary habits. We also introduce accompanying metrics. The method generates and compares word embeddings to identify the parent food category of each food entry and then calculates the most popular. Our proposed approach identifies 82% of a user's ten most frequently eaten foods. Our method is publicly available on (https://github.com/aametwally/LearningFoodPreferences)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge